Molecular characterisation of methicillinsensitive Staphylococcus aureus from deep






- Slides: 17

Molecular characterisation of methicillin-sensitive Staphylococcus aureus from deep surgical site infections in orthopaedic patients

Background • Staphylococcus aureus is a leading cause of surgical site infections (SSIs) , which are found in more than 50% of infections related to total joint arthroplasty. • However, a recent study showed that nasal S. aureus carriage did not predict S. aureus surgical site infections(SSI).

Background • The aim of the study was to identify bacterial molecular markers that distinguish S. aureus strains causing deep SSI from nasal carrier strains in patients undergoing major orthopaedic surgery. • In order to cover a large portion of the involved S. aureus genes, a DNA microarray covering approximately 170 bacterial genes and their allelic variants was used to characterise the strains.

Materials and methods • A total of 117 S. aureus isolates obtained from patients undergoing major orthopaedic surgery in 2004 -2006 were included. Isolates from 60 patients with deep SSI were collected by swabs from secreting wounds or intraoperative biopsies at soft tissue revision. • As controls, 57 nasal S. aureus isolates were collected pre-operatively from patients admitted to elective orthopaedic procedures.

Clinical information • Patients were categorised into two groups according to the ASA score (<2 and >2). Deep SSI was defined using the Centers for Disease Control and Prevention (CDC) criteria.

Results • • Clinical data Clonal complexes Accessory regulatory gene (agr) groups Microbial surface components recognising adhesive matrix molecules (MSCRAMM) genes Capsule types Virulence factors β-hemolysin converting phages Resistance genes

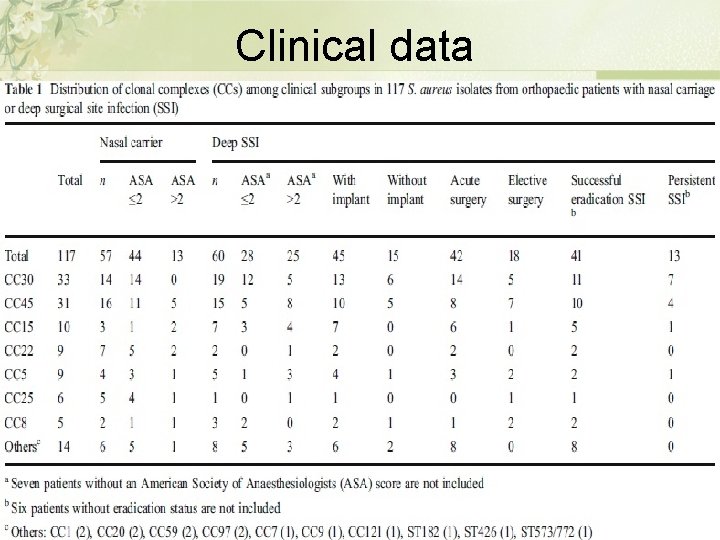
Clinical data

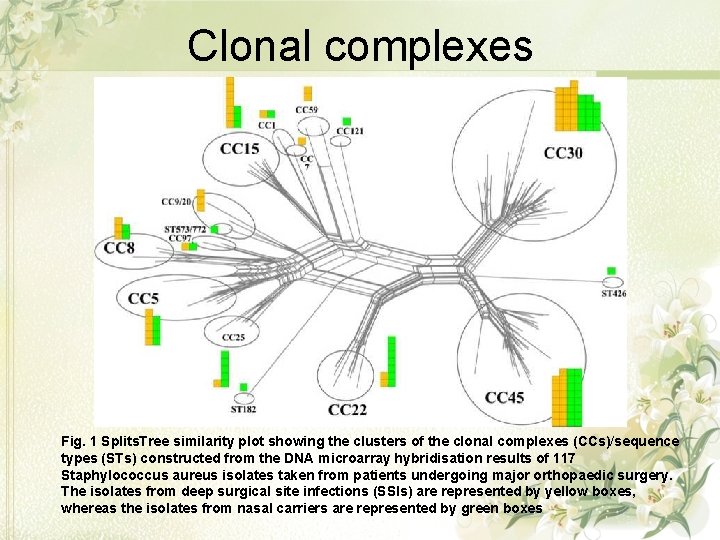
Clonal complexes Fig. 1 Splits. Tree similarity plot showing the clusters of the clonal complexes (CCs)/sequence types (STs) constructed from the DNA microarray hybridisation results of 117 Staphylococcus aureus isolates taken from patients undergoing major orthopaedic surgery. The isolates from deep surgical site infections (SSIs) are represented by yellow boxes, whereas the isolates from nasal carriers are represented by green boxes

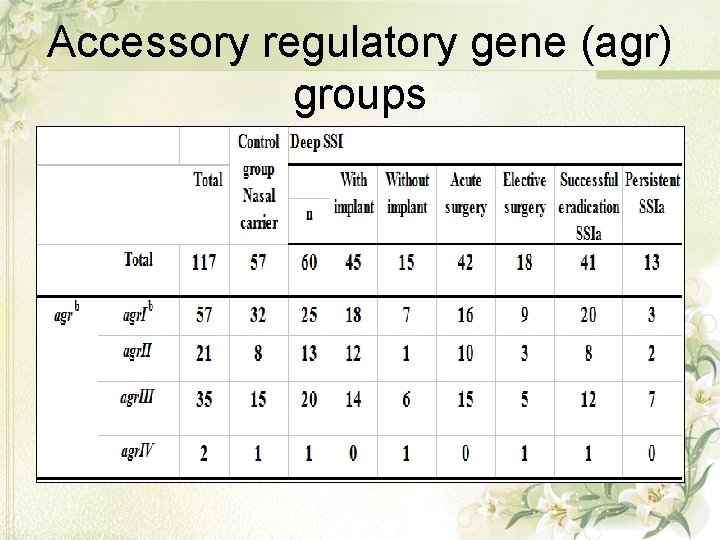
Accessory regulatory gene (agr) groups

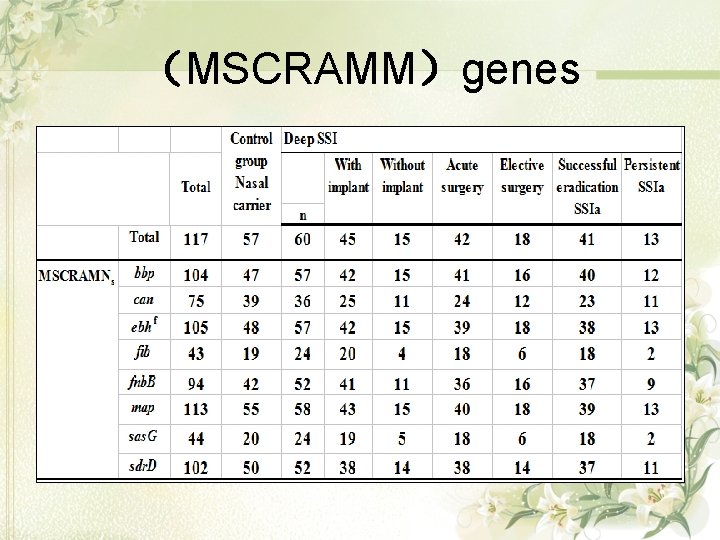
(MSCRAMM)genes

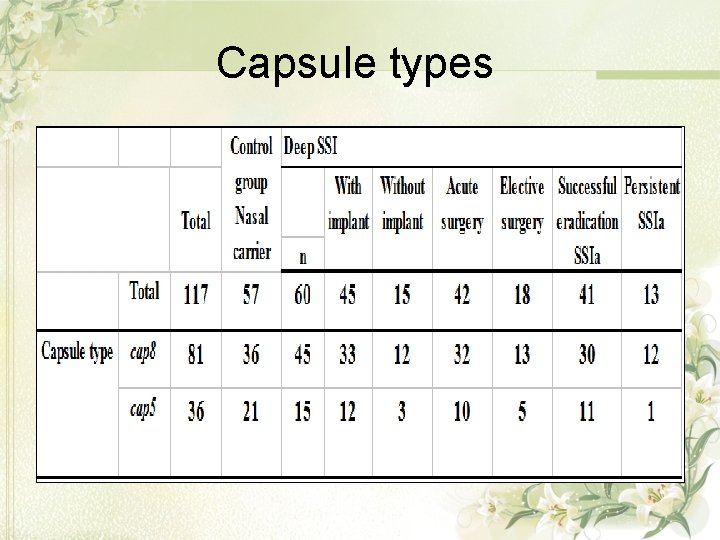
Capsule types

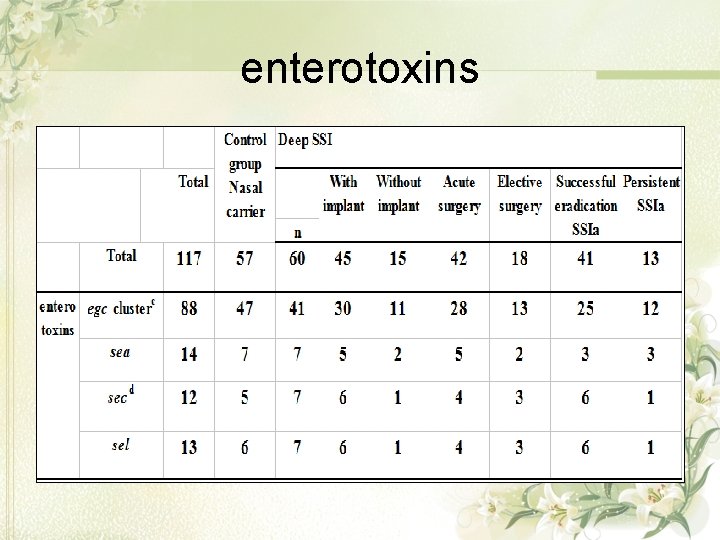
enterotoxins

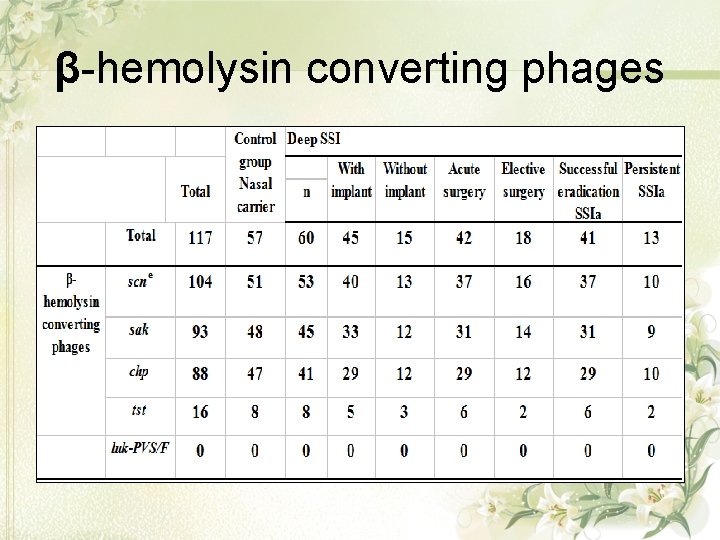
β-hemolysin converting phages

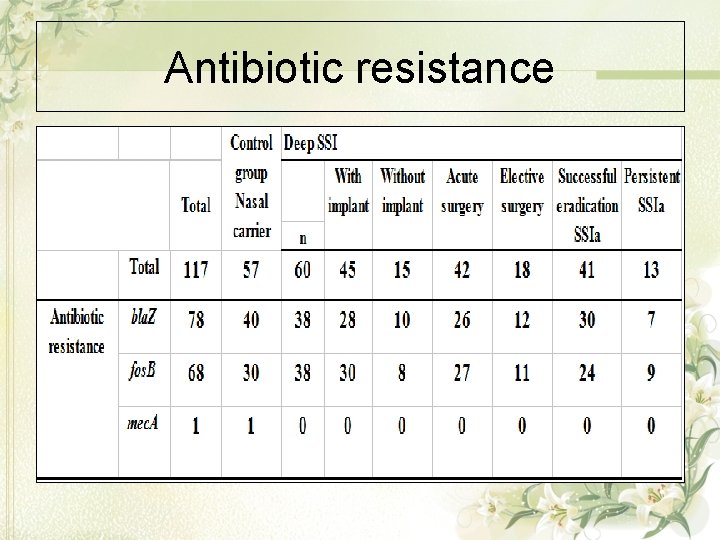
Antibiotic resistance

Discussion • The MSCRAMM gene bbp was more frequent in deep SSI than in nasal carrier isolates, suggesting a role in S. aureus invasiveness in deep SSI. • Agr, thereby, plays an important role in S. aureus pathogenesis. • The presence of sak might be linked to higher invasiveness of S. aureus, finding the gene more frequently in patients than in asymptomatic carriers.

Discussion • Our study contributes new genotypic information on methicillin-sensitive S. aureus isolates from deep SSI in orthopaedic patients. • Further studies to unravel the interface between S. aureus, host infection susceptibility and innate immunity are important for a more extensive understanding of the pathogenesis of SSIs.

Thank you!