Molecular biology Transformation introduction of DNA Selectable marker
Molecular biology • Transformation: introduction of DNA – Selectable marker – Spheroplasts, Li 2+ salts, electroporation • Yeast plasmids are shuttle plasmids, amplification in E. coli, –ori, b-lactamase • Transformation with oligonucleotides, -> selection • Transformation of mitochondria by particle bombardment

Important genes • URA 3, LYS 2, can be negative selected against (FOA, aaminoadipic acid) – Host alleles, ura 3 -52 -> ura 3∆0 • Dominant drug selectable markers, i. e. kan. MX • ADE 1, ADE 2, ade 1 and ade 2 mutants produce a red pigment – But ade 3 ade 2 is white – Colony sectoring screen – Syntehtic lethal secreen • GAL promoter for conditional expression • lac. Z, GFP fusions • Epitope tags
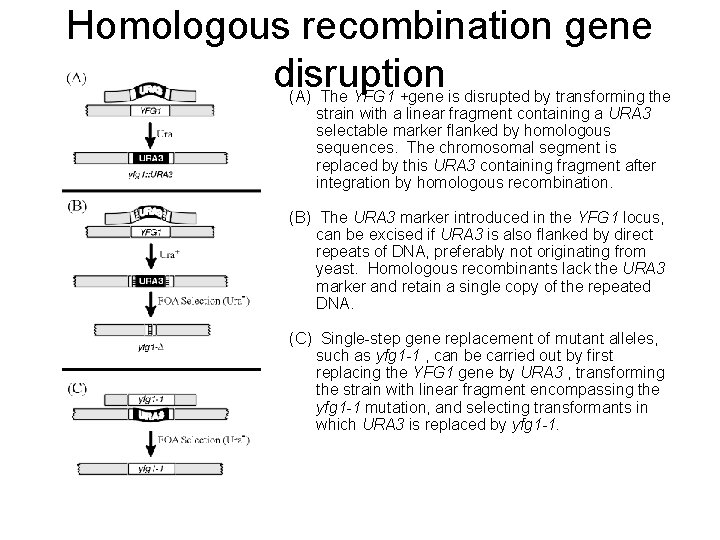
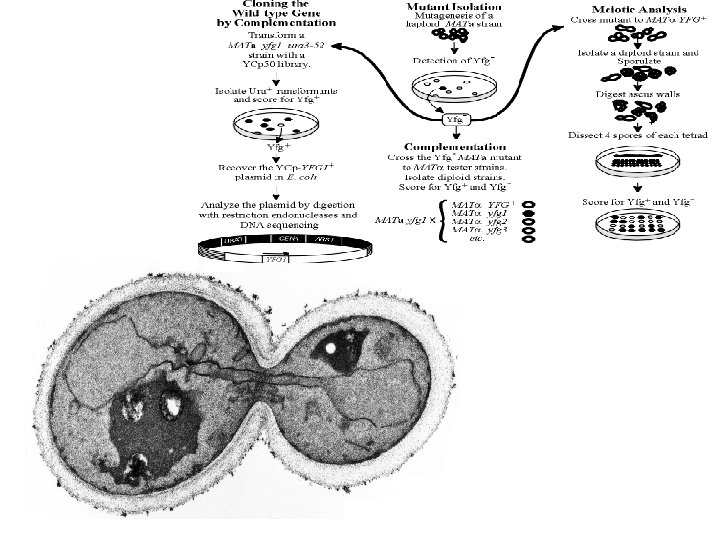
Homologous recombination gene disruption (A) The YFG 1 +gene is disrupted by transforming the strain with a linear fragment containing a URA 3 selectable marker flanked by homologous sequences. The chromosomal segment is replaced by this URA 3 containing fragment after integration by homologous recombination. (B) The URA 3 marker introduced in the YFG 1 locus, can be excised if URA 3 is also flanked by direct repeats of DNA, preferably not originating from yeast. Homologous recombinants lack the URA 3 marker and retain a single copy of the repeated DNA. (C) Single-step gene replacement of mutant alleles, such as yfg 1 -1 , can be carried out by first replacing the YFG 1 gene by URA 3 , transforming the strain with linear fragment encompassing the yfg 1 -1 mutation, and selecting transformants in which URA 3 is replaced by yfg 1 -1.
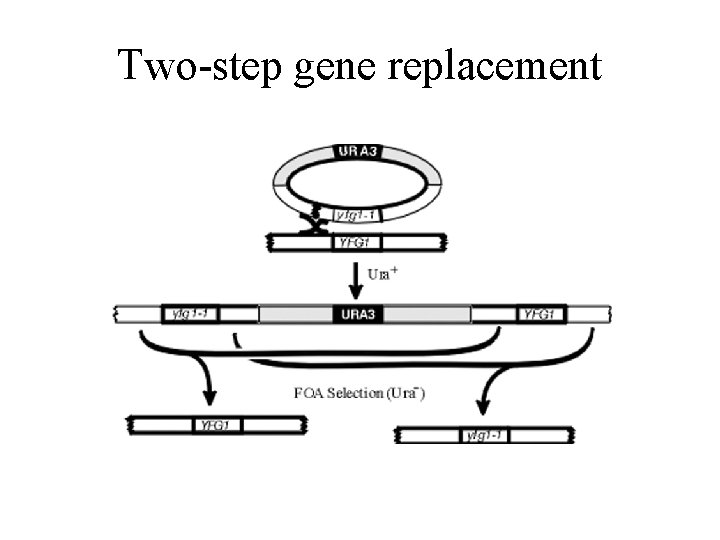
Two-step gene replacement

4 types of yeast vectors
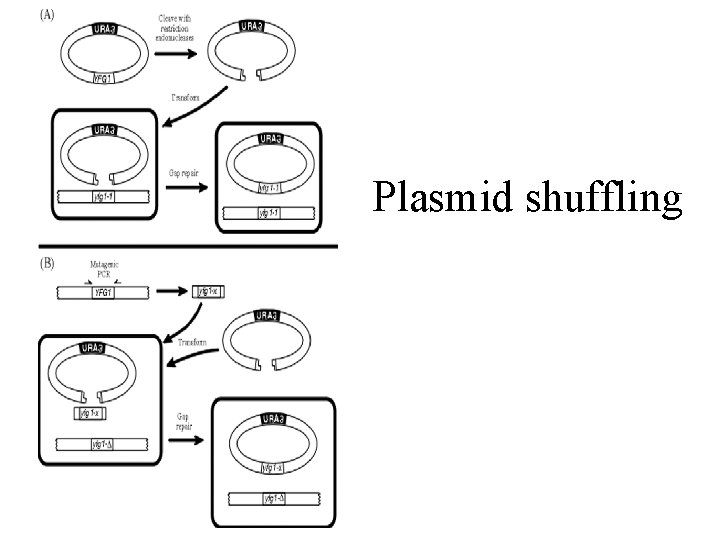
Plasmid shuffling
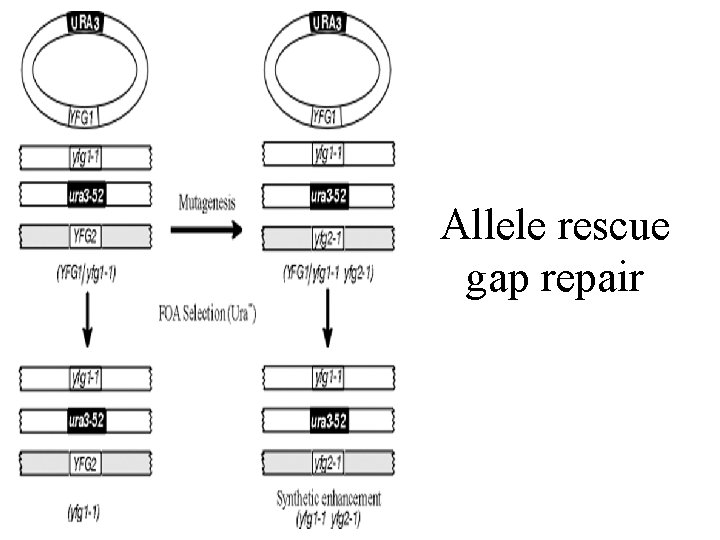
Allele rescue gap repair
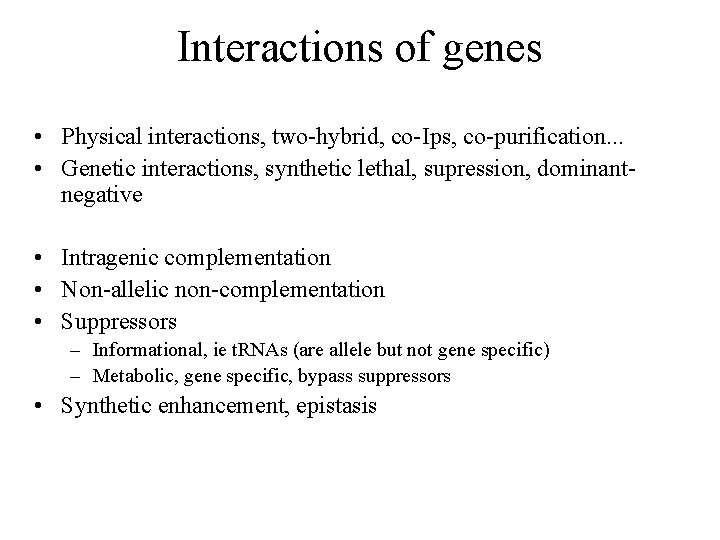
Interactions of genes • Physical interactions, two-hybrid, co-Ips, co-purification. . . • Genetic interactions, synthetic lethal, supression, dominantnegative • Intragenic complementation • Non-allelic non-complementation • Suppressors – Informational, ie t. RNAs (are allele but not gene specific) – Metabolic, gene specific, bypass suppressors • Synthetic enhancement, epistasis
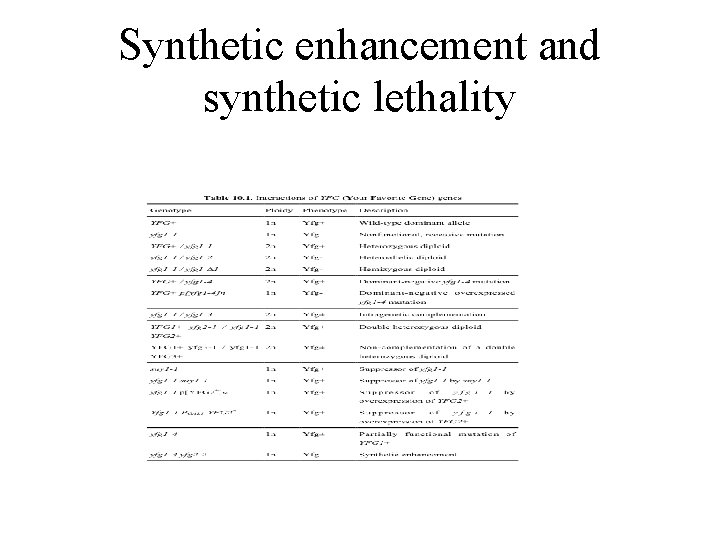
Synthetic enhancement and synthetic lethality
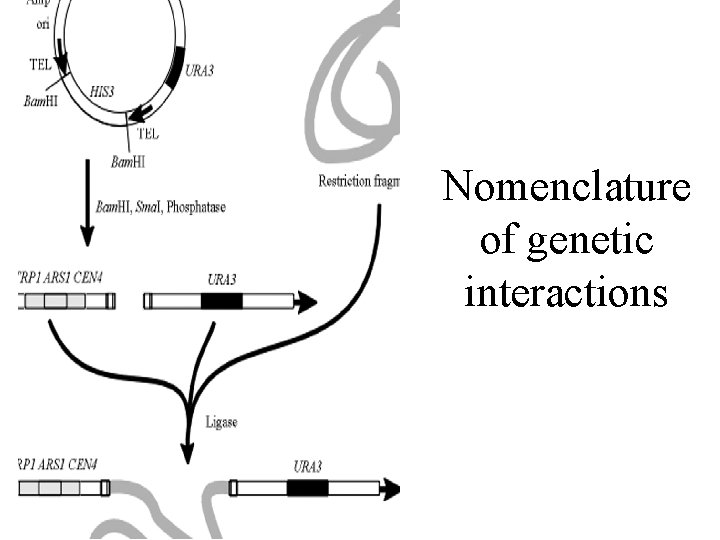
Nomenclature of genetic interactions
Reverse genetics • Gene -> phenotype -> function (annotation)
Yeast specific Methods • Two hybrid • Yeast Artifical Chromosomes (YACs) (50 - 500 kb) • Expression of heterologous proteins – No endotoxins – Posttranslational modifications (acetylation, myristoylation, . . ) – secretion
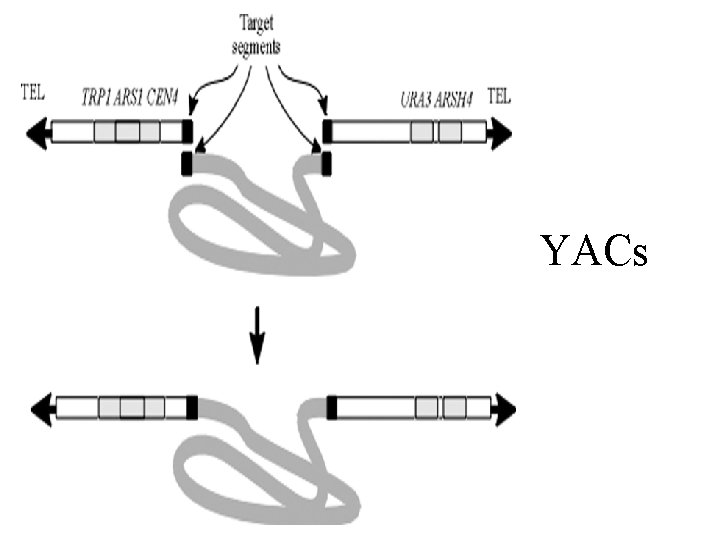
YACs
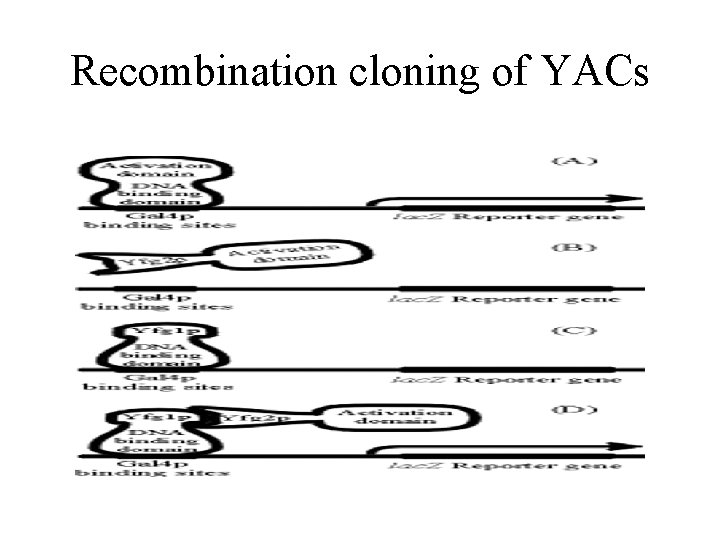
Recombination cloning of YACs
The two-hybrid system
Protein interaction map
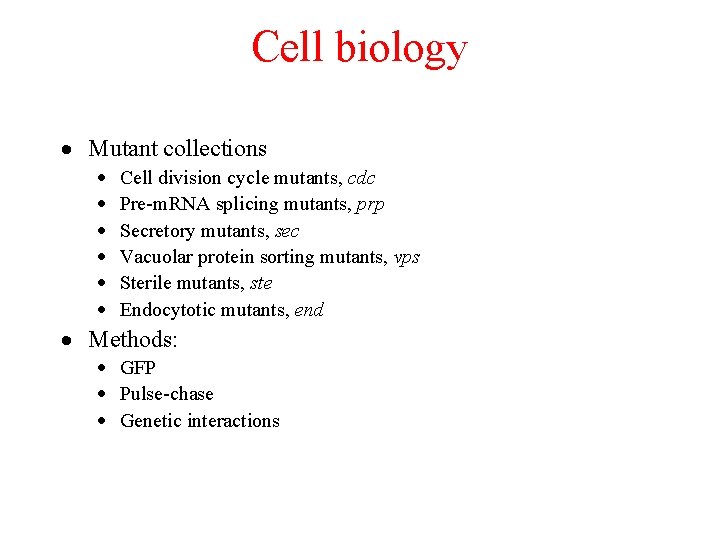
Cell biology · Mutant collections · · · Cell division cycle mutants, cdc Pre-m. RNA splicing mutants, prp Secretory mutants, sec Vacuolar protein sorting mutants, vps Sterile mutants, ste Endocytotic mutants, end · Methods: · GFP · Pulse-chase · Genetic interactions
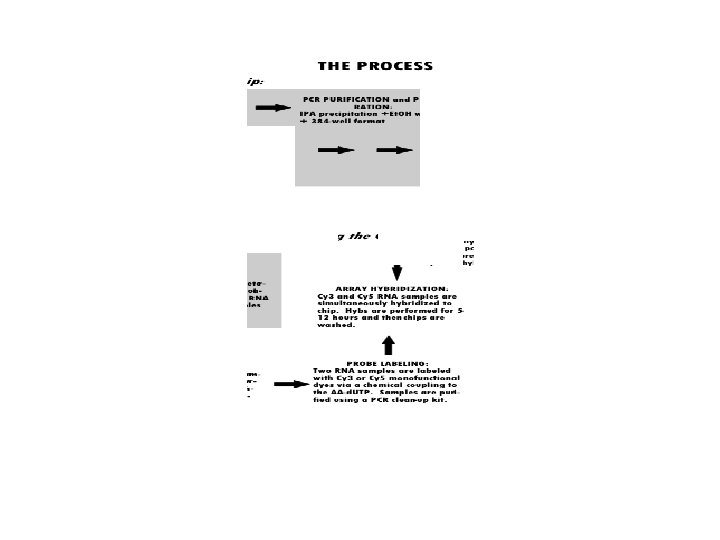
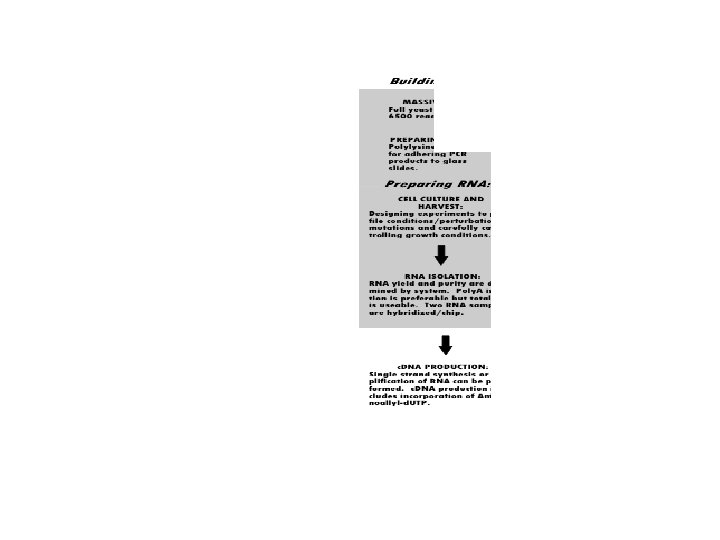

Gene-omics • Microarrays (DNA, oligo) • comparative, yeast (sensus stricto), . . . • Proteomics, MS, chips (120 kinases) • Metabolomics, flux of metabolites
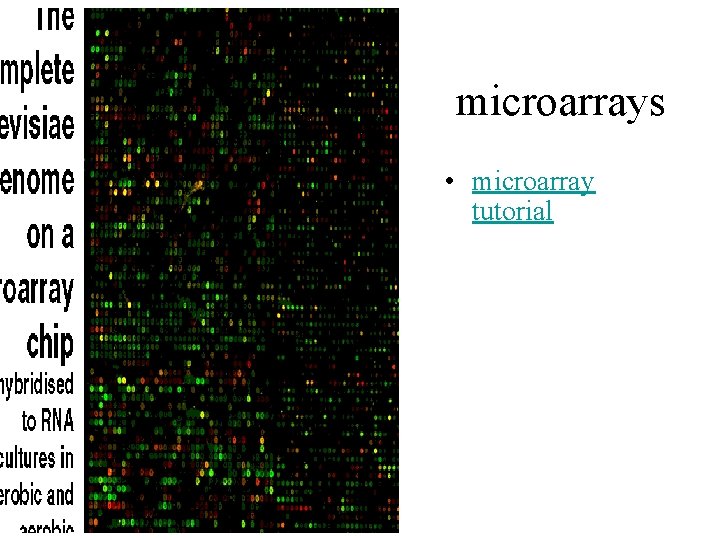
microarrays • microarray tutorial
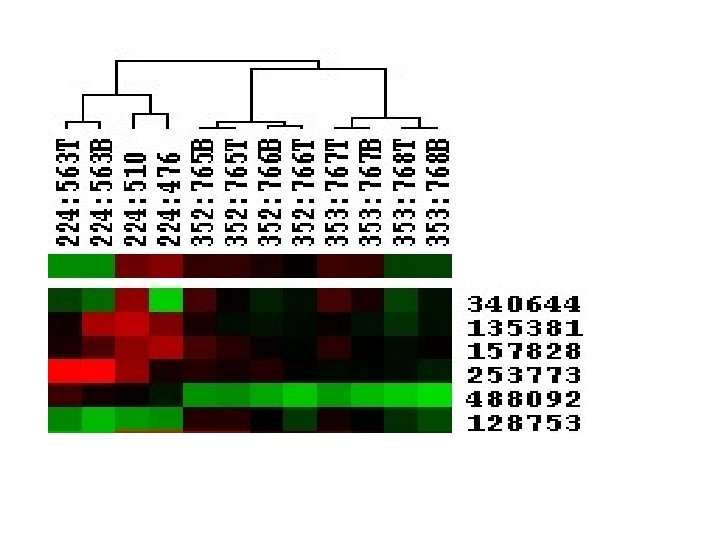

clustering

SAGE • • ACO T 56 Cell 1997, 243 Transcripts 0. 3 - 200 / cell Only 18% of genes have > 100 transcripts (energy metab. ribosome) • No transcript clustering in genome
- Slides: 25