Molecular Biology PROTEIN SYNTHESIS Proteins DNA codes for
Molecular Biology PROTEIN SYNTHESIS
Proteins • • DNA codes for proteins Proteins provide phenotypic traits Genotype – genetic make up Phenotype – physical characteristics or specific traits • Genes are found on chromosomes • The genes provide the instructions for making proteins • DNA RNA Protein Synthesis
• There are 2 steps to protein synthesis ▫ Transcription – the transfer of genetic information from DNA into a RNA molecule ▫ Translation – transfer of RNA into a protein
DNA Nucleus Cytoplasm
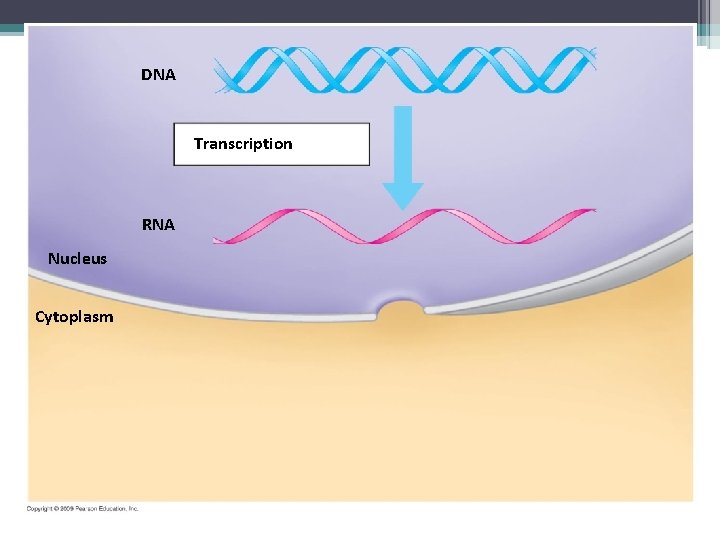
DNA Transcription RNA Nucleus Cytoplasm
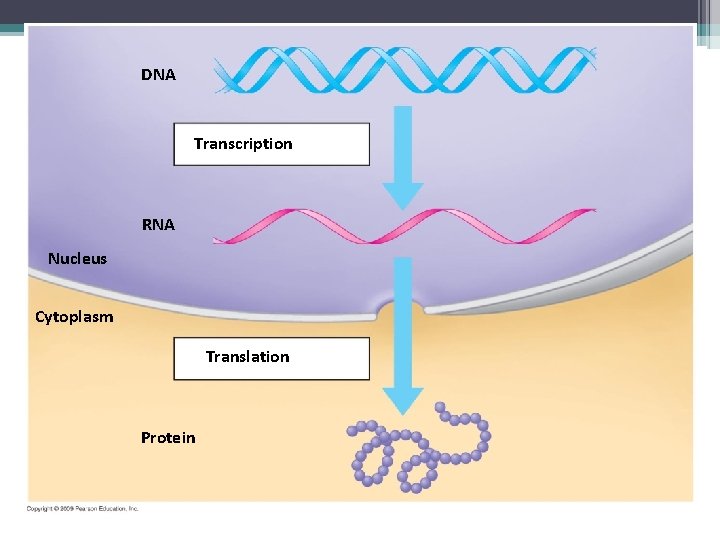
DNA Transcription RNA Nucleus Cytoplasm Translation Protein

Archibold Garrod • 1909 • Suggested genes dictate phenotypes through enzymes • He studied inherited diseases • He said diseases reflected a person’s inability to make an enzyme • Ex – alkaptonuria causes urine to be black because of the absence of an enzyme to break down alkapton

Beadle and Tatum • 1940 s • They worked with bread mold – Neurospora crassa • They studied strains of mold that could not grow on a simple growth medium • Each of these nutritional mutants lacked an enzyme in a metabolic pathway that make some molecule the mold needed (amino acid) • Showed each mutant was defective in a single gene • This led to the 1 gene – 1 enzyme hypothesis

One gene – One polypeptide Hypothesis • One gene –one enzyme became one gene – one protein hypothesis ▫ This is because not all proteins are enzymes • This hypothesis soon became one gene – one polypeptide ▫ This is because many proteins are made up of two or more polypeptide chains

Codons • The sequence of nucleotides in DNA provides a code for constructing a protein • Protein construction requires a conversion of a nucleotide sequence to an amino acid sequence • The information comes from genes • Genes - section of DNA that is made up of hundreds or thousands of nucleotides in a specific order

Codons • Each amino acid is specified by a codon • 1 codon is made up of 3 nucleotides • 64 codons are possible • Some amino acids have more than one possible codon • CAGAUUUAUCCG – How many codons are here?

Transcription and Translation • Transcription – a gene is copied and a complementary RNA strand is formed Remember there is no thymine in RNA instead there is URACIL Rewrites the DNA code into RNA, using the same nucleotide “language” • Translation – converts the RNA strand into a protein Remember that amino acids are the monomers of proteins Switching from the nucleotide “language” to amino acid “language”
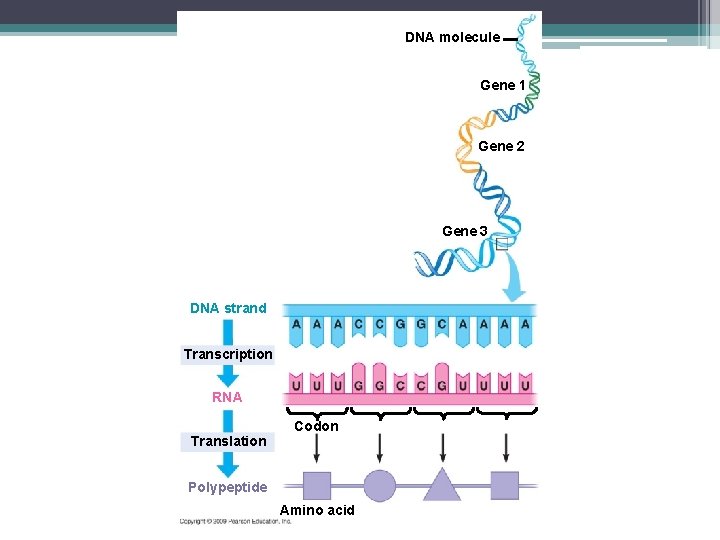
DNA molecule Gene 1 Gene 2 Gene 3 DNA strand Transcription RNA Codon Translation Polypeptide Amino acid

DNA strand Transcription RNA Codon Translation Polypeptide Amino acid

Genetic Codes • Genetic code – the set of rules giving the information between codons in RNA and amino acids of proteins • Three nucleotides specify one amino acid • 61 codons correspond to amino acids – AUG codes for methionine and signals the start of transcription – 3 “stop” codons signal the end of translation (UAA, UAG, UGA) Redundant: More than one codon for some amino acids

Genetic Codes – “Wobble position” – the first two bases count more than the third amino acid in a codon. – Nearly universal so evolved early on in life

Third base First base Second base

Strand to be transcribed DNA

Strand to be transcribed DNA Transcription RNA Start codon Stop codon
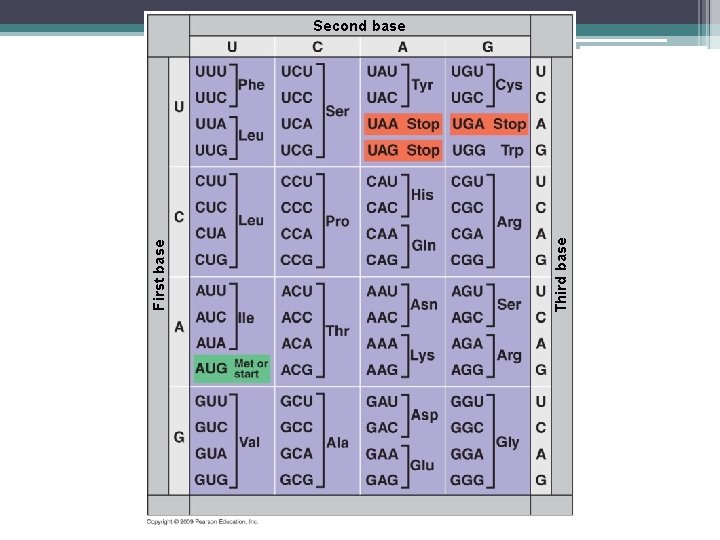
Third base First base Second base

Strand to be transcribed DNA Transcription RNA Start codon Polypeptide Met Translation Lys Phe Stop codon

Transcription • Happens in the nucleus of eukaryotic cells • An RNA molecule is made from DNA because DNA cannot leave the nucleus • The DNA strands separate but only one acts as a template • Complementary RNA nucleotides to the DNA template strand are added to form the m. RNA strand

Transcription • RNA polymerase – enzyme that helps add RNA nucleotides • Remember there is no thymine in RNA • The Promoter ▫ A DNA sequence has a “start transcribing” signal made up of a specific nucleotide sequence ▫ RNA polymerase binds to the promoter ▫ The promoter also tells which DNA strand is the template

Steps of Transcription • Initiation ▫ Attachment of RNA polymerase to promoter ▫ Start of RNA synthesis • Elongation ▫ RNA nucleotides continue to be attached ▫ The two strands which are being held together by H bonds start to separate and the DNA molecules retwist

Steps of Transcription • Termination ▫ The RNA polymerase reaches the terminator ▫ Terminator – a sequence that signals the end of the gene ▫ RNA polymerase detaches.
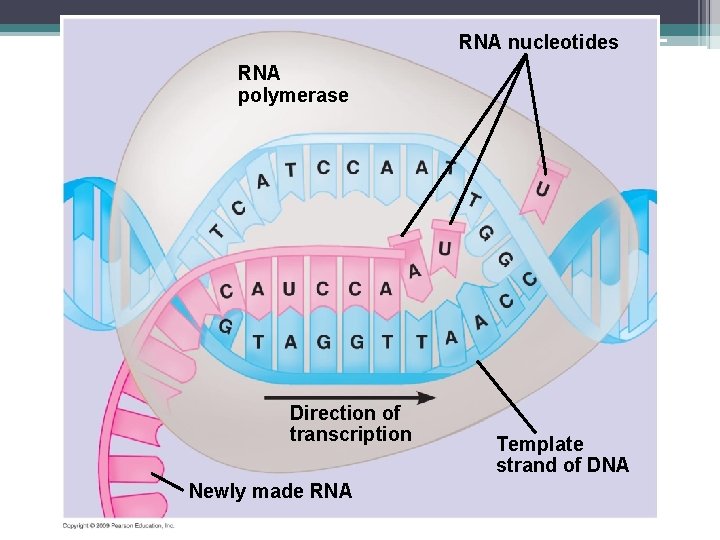
RNA nucleotides RNA polymerase Direction of transcription Newly made RNA Template strand of DNA

RNA polymerase DNA of gene Promoter DNA Terminator DNA 1 Initiation 2 Elongation 3 Termination Completed RNA Area shown in Figure 10. 9 A Growing RNA polymerase
Modification of m. RNA • Messenger RNA (m. RNA) contains codons for protein sequences • The m. RNA strand is processed before it leaves the nucleus through a nuclear pore • A cap (5’ end: single guanine nucleotide) and a tail added to (3’ end: Poly-A tail of 50– 250 adenines) is added to the m. RNA strand – This helps with the export of m. RNA – Protects the m. RNA from being degraded by enzymes – Helps the m. RNA strand to bind to the ribosomes

Modification of m. RNA • Introns have to be removed ▫ Genes do have some noncoding areas ▫ These noncoding sequences are called introns ▫ Exons – are the coding regions of the gene These are the parts of the gene that are expressed as amino acids ▫ Introns get removed and exons get joined together ▫ This cutting and pasting is called RNA splicing
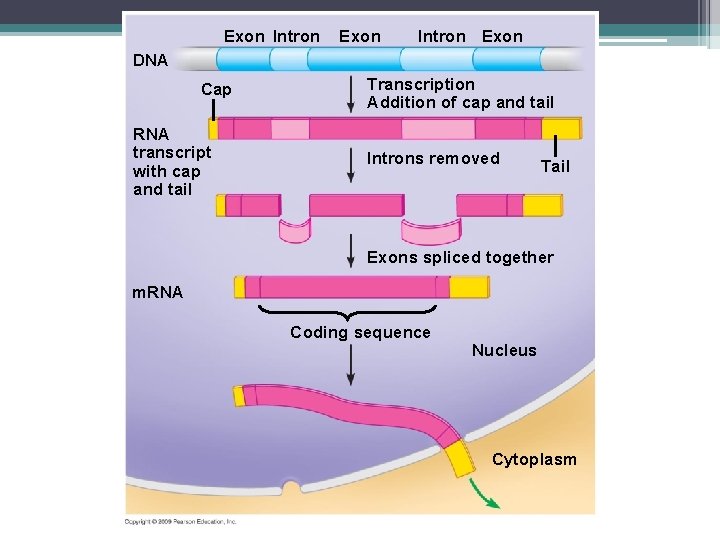
Exon Intron Exon DNA Cap RNA transcript with cap and tail Transcription Addition of cap and tail Introns removed Tail Exons spliced together m. RNA Coding sequence Nucleus Cytoplasm

Transfer RNA (t. RNA) • Helps to read m. RNA to make a protein • Amino acids are found in the cytoplasm • t. RNAs help to transfer these amino acids to help make proteins • t. RNA has to match the amino acid to the codon on the m. RNA strand
Transfer RNA (t. RNA) • Structure ▫ Made up of a single strand of RNA made up of about 80 nucleotides ▫ Folds and twists on itself ▫ At one end is the anticodon – special triplet of bases that is complementary to a codon on the m. RNA strand ▫ The other end has the site for an amino acid to attach ▫ There is an enzyme here that makes sure to join the correct amino acid to the t. RNA molecule
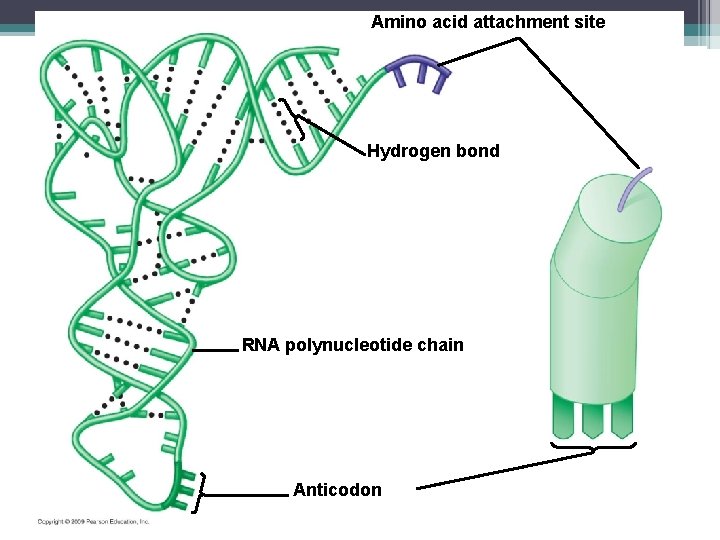
Amino acid attachment site Hydrogen bond RNA polynucleotide chain Anticodon


Ribosomes • • • Helps with the making of the protein They are found in the cytoplasm Made up of 2 subunits Made up of ribosomal RNA (r. RNA) They have a binding site for m. RNA and two binding sites or t. RNA
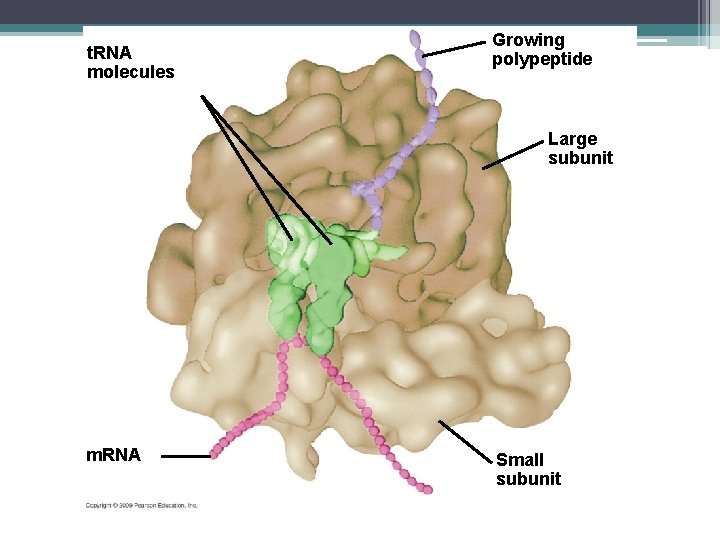
t. RNA molecules Growing polypeptide Large subunit m. RNA Small subunit
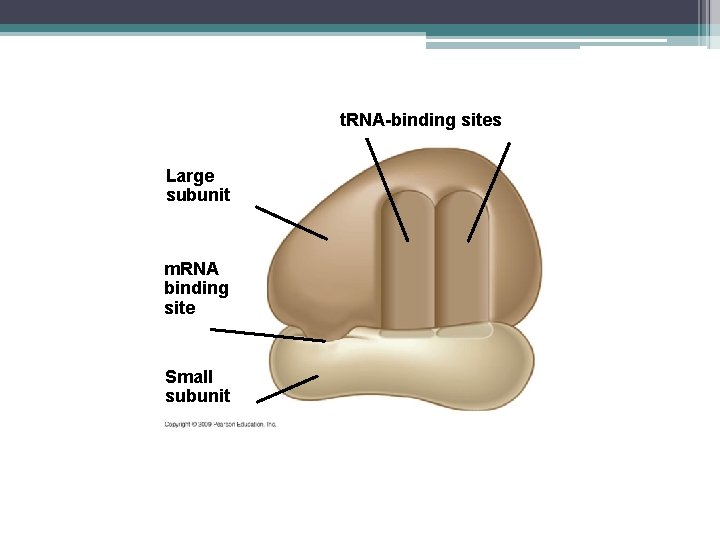
t. RNA-binding sites Large subunit m. RNA binding site Small subunit

Next amino acid to be added to polypeptide Growing polypeptide t. RNA m. RNA Codons
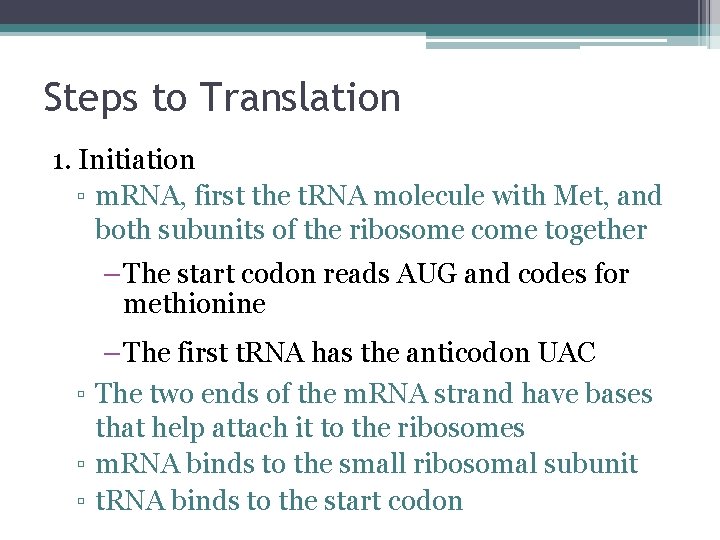
Steps to Translation 1. Initiation ▫ m. RNA, first the t. RNA molecule with Met, and both subunits of the ribosome come together – The start codon reads AUG and codes for methionine – The first t. RNA has the anticodon UAC ▫ The two ends of the m. RNA strand have bases that help attach it to the ribosomes ▫ m. RNA binds to the small ribosomal subunit ▫ t. RNA binds to the start codon
Start of genetic message End

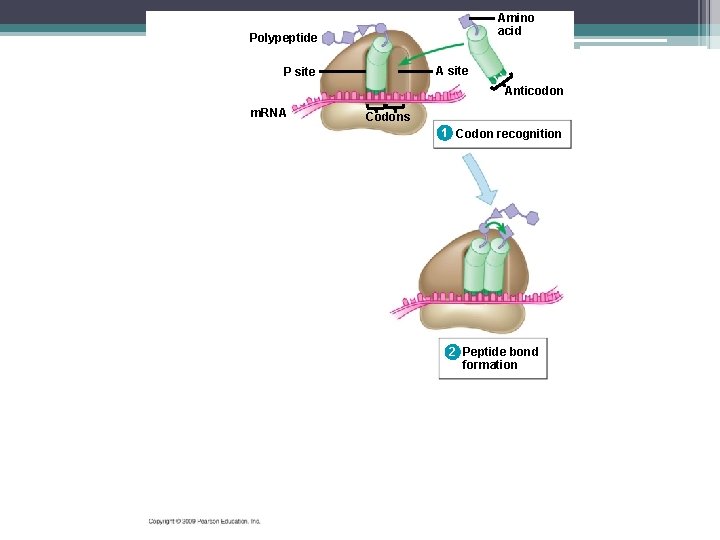
Steps to Translation ▫ The large ribosomal subunit binds to the small subunit ▫ The first t. RNA is in one of the two t. RNA binding sites on the ribosomes ▫ This is the P site. ▫ P site – holds the growing polypeptide chain ▫ A site – is vacant and is waiting for the next t. RNA molecule

Met Initiator t. RNA P site 1 m. RNA Start codon Small ribosomal subunit 2 Large ribosomal subunit A site

Steps to Translation 2. Elongation – Amino acids are added one by one to the first amino acid (Met) – a 3 step process: 1. Codon recognition: next t. RNA’s anticodon binds to the complementary codon on the m. RNA at the A site 2. Peptide bond formation: joining of the new amino acid to the chain –Amino acids on the t. RNA at the P site are attached by a covalent bond to the amino acid on the t. RNA at the A site –The ribosome helps with the bonding

Steps to Translation Elongation (Con’t) 3. Translocation - t. RNA is released from the P site and the ribosome moves t. RNA from the A site into the P site ▫ So the next codon on the m. RNA is in the A site.

Steps to Translation 3. Termination ▫ Elongation continues until a stop codon (UAA, UAG, UGA) ▫ A release factor adds on a water instead of an amino acid ▫ Everything breaks apart

Amino acid Polypeptide A site P site Anticodon m. RNA Codons 1 Codon recognition
Amino acid Polypeptide A site P site Anticodon m. RNA Codons 1 Codon recognition 2 Peptide bond formation
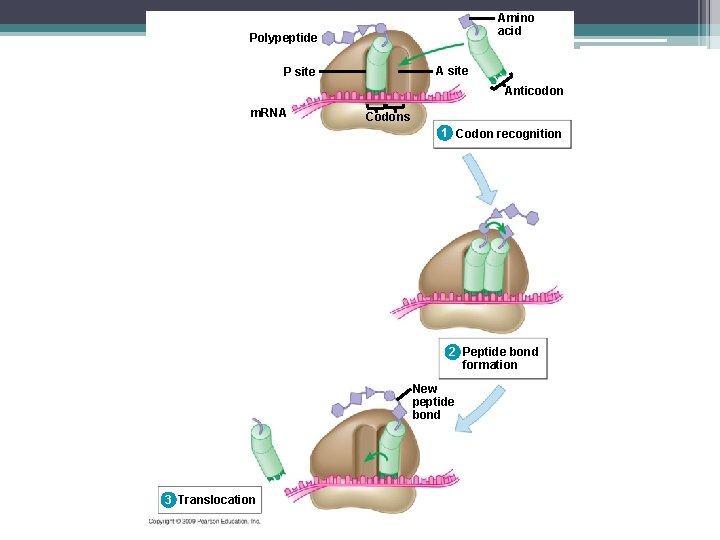
Amino acid Polypeptide A site P site Anticodon m. RNA Codons 1 Codon recognition 2 Peptide bond formation New peptide bond 3 Translocation
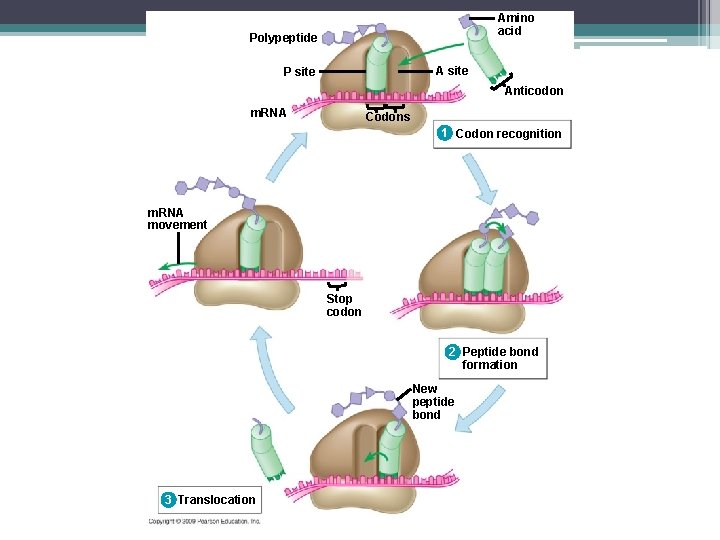
Amino acid Polypeptide A site P site Anticodon m. RNA Codons 1 Codon recognition m. RNA movement Stop codon 2 Peptide bond formation New peptide bond 3 Translocation

Review ▫ Does translation represent: – DNA RNA or RNA protein? ▫ Where does the information for producing a protein originate: – DNA or RNA? ▫ Which one has a linear sequence of codons: – r. RNA, m. RNA, or t. RNA? ▫ Which one directly influences the phenotype: – DNA, RNA, or protein?

Transcription DNA m. RNA Amino acid RNA polymerase 1 m. RNA is transcribed from a DNA template. Translation 2 Each amino acid attaches to its proper t. RNA with the help of a specific enzyme and ATP. Enzyme ATP t. RNA Anticodon Large ribosomal subunit Initiator t. RNA 3 Initiation of polypeptide synthesis The m. RNA, the first t. RNA, and the ribosomal sub-units come together. Start Codon m. RNA Small ribosomal subunit
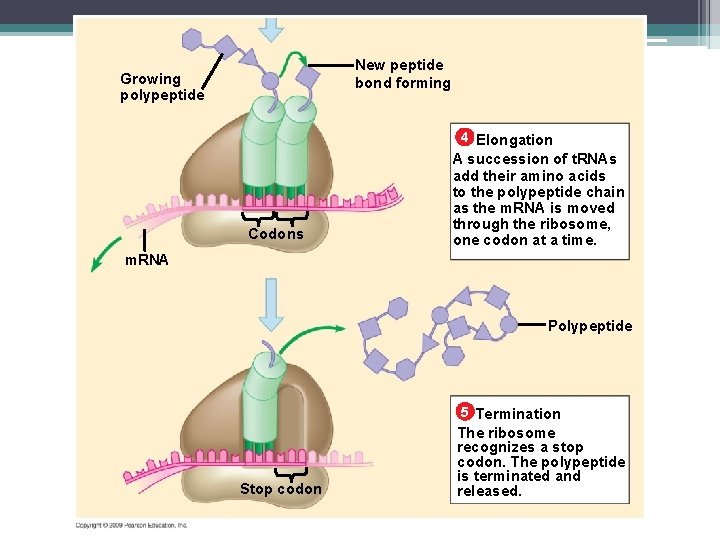
New peptide bond forming Growing polypeptide 4 Elongation Codons A succession of t. RNAs add their amino acids to the polypeptide chain as the m. RNA is moved through the ribosome, one codon at a time. m. RNA Polypeptide 5 Termination Stop codon The ribosome recognizes a stop codon. The polypeptide is terminated and released.
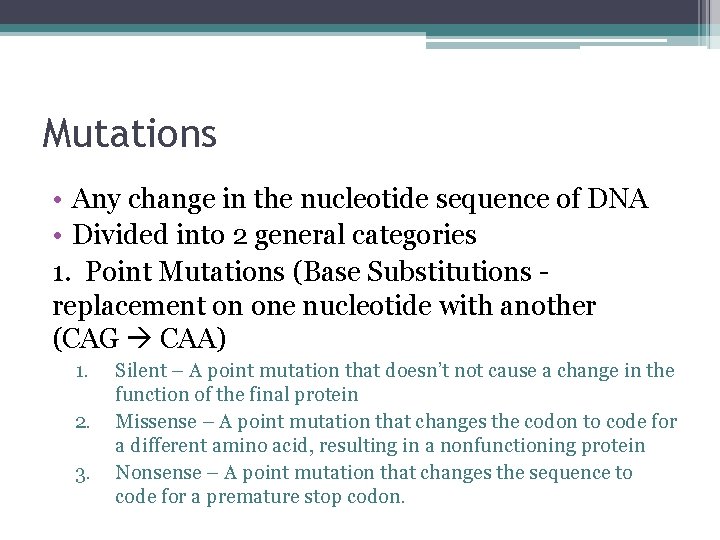
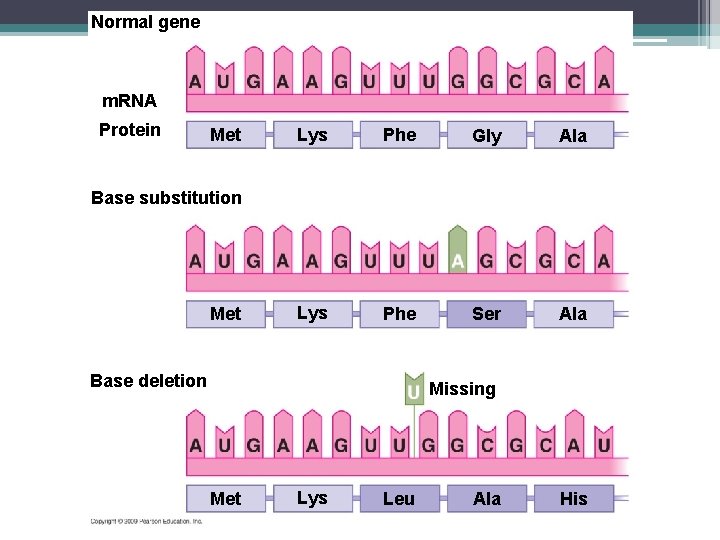
Mutations • Any change in the nucleotide sequence of DNA • Divided into 2 general categories 1. Point Mutations (Base Substitutions replacement on one nucleotide with another (CAG CAA) 1. 2. 3. Silent – A point mutation that doesn’t not cause a change in the function of the final protein Missense – A point mutation that changes the codon to code for a different amino acid, resulting in a nonfunctioning protein Nonsense – A point mutation that changes the sequence to code for a premature stop codon.
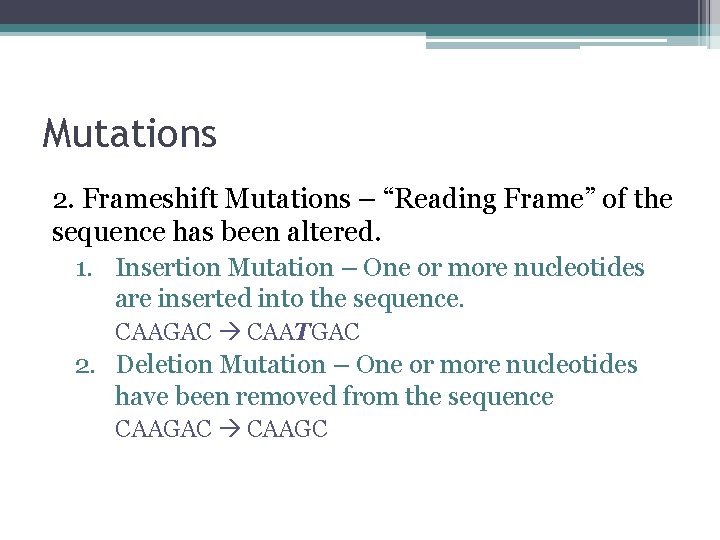
Mutations 2. Frameshift Mutations – “Reading Frame” of the sequence has been altered. 1. Insertion Mutation – One or more nucleotides are inserted into the sequence. CAAGAC CAATGAC 2. Deletion Mutation – One or more nucleotides have been removed from the sequence CAAGAC CAAGC
Mutations • Duplication Mutation – A region of a chromosome is copied, resulting in multiple copies of that region • Inversion Mutation –The order of a segment of chromosome is reversed. • Mutagenesis – production of mutations ▫ Can be spontaneous during DNA replication ▫ Can also result from mutagens – physical or chemical agent that can cause mutations (ex- radiation) Carcinogen – A mutagen that causes cancer
Normal gene m. RNA Protein Met Lys Phe Gly Ala Lys Phe Ser Ala Base substitution Met Base deletion Missing Met Lys Leu Ala His
- Slides: 57