Modular Biocatalyst Platform for Chiral Synthesis of Chemical

Modular Biocatalyst Platform for Chiral Synthesis of Chemical Compounds by Structure-based Directed Evolution the BIOCAT project Marinus G. Casteleijn 10 -11 February, Helsinki KETJU meeting
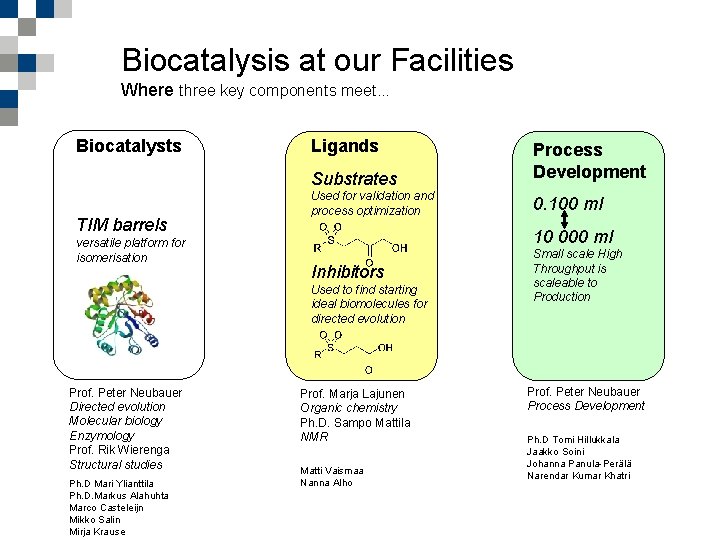
Biocatalysis at our Facilities Where three key components meet. . . Biocatalysts Ligands Substrates TIM barrels versatile platform for isomerisation Used for validation and process optimization Ph. D Mari Ylianttila Ph. D. Markus Alahuhta Marinus G. Casteleijn Marco Casteleijn 10 -11 February, Helsinki Mikko Salin KETJU meeting Mirja Krause 0. 100 ml 10 000 ml Inhibitors Used to find starting ideal biomolecules for directed evolution Prof. Peter Neubauer Directed evolution Molecular biology Enzymology Prof. Rik Wierenga Structural studies Process Development Prof. Marja Lajunen Organic chemistry Ph. D. Sampo Mattila NMR Matti Vaismaa Nanna Alho Small scale High Throughput is scaleable to Production Prof. Peter Neubauer Process Development Ph. D Tomi Hillukkala Jaakko Soini Johanna Panula-Perälä Narendar Kumar Khatri
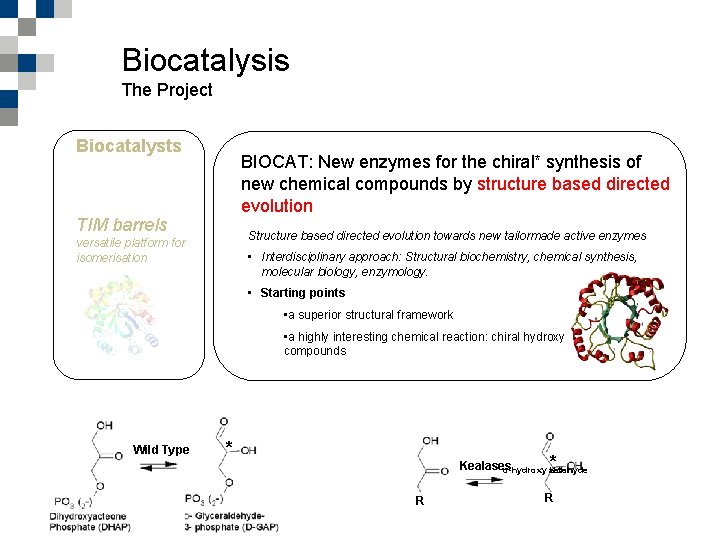
Biocatalysis The Project Biocatalysts BIOCAT: New enzymes for the chiral* synthesis of new chemical compounds by structure based directed evolution TIM barrels Structure based directed evolution towards new tailormade active enzymes versatile platform for isomerisation • Interdisciplinary approach: Structural biochemistry, chemical synthesis, molecular biology, enzymology. • Starting points • a superior structural framework • a highly interesting chemical reaction: chiral hydroxy compounds Wild Type Marinus G. Casteleijn 10 -11 February, Helsinki KETJU meeting * * Kealases α-hydroxy aldehyde keton R R

Proof-Of-Principle studies Characterization of monomeric TIMs Binding studies NMR/Mass Spectrometry Chemical synthesis X-ray/docking A-TIM-A 178 L A-TIM-S 96 P A-TIM-I 245 A Start Directed Evolution A-TIM-X* Directed Evolution A-TIM-Y** Screening Selection *Rpi. A/B activity *Ara. A activity *Xyl. A activity Added based on the previous KETJU meeting Marinus G. Casteleijn 10 -11 February, Helsinki KETJU meeting Active enzymes **new activity

The libraries – selection of good targets A 178 L Lead enzyme S 96 P ATIM Rational Design: Site-directed mutagenesis creates four starting points for the directed evolution approach I 245 A Starting points (4) 4 Starting points - ATIM (A) - ATIM-S 96 P (ASP) - ATIM-A 178 L (AAL) Marinus G. Casteleijn 10 -11 February, Helsinki KETJU meeting - ATIM-I 245 A (AIA) ATIM (A) ATIM-S 96 P (ASP) ATIM-A 178 L (AAL) ATIM-I 245 A (AIA)

The libraries – selection of good targets Mutagenesis targeted random W 100 V 214/ N 215 Rational Design: Megaprimer PCR creates different libraries of ATIM mutants Regions (3) Targeted mutagenesis (megaprimer method ) W 100 (W) 3 Regions A 233/G 234/ V 214/N 215 (VN) - W 100 (W) K 239/E 241 A 233/G 234/K 239/E 2 41 - V 214/N 215 (VN) - A 233/G 234/K 239/E 241 (AGKE) Marinus G. Casteleijn 10 -11 February, Helsinki KETJU meeting (AGKE)

The libraries – creating the experimental space 16 libraries of A-TIM variants Fully randomized mutagenesis Starting points (4) - ATIM (A) - ATIM-S 96 P (ASP) - ATIM-A 178 L (AAL) - ATIM-I 245 A (AIA) Marinus G. Casteleijn 10 -11 February, Helsinki KETJU meeting Error rate 0. 3– 0. 6 % amino acid change (Fu) Targeted mutagenesis (megaprimer method ) Results Regions (3) Libraries (16) - W 100 (W) - A (Fu, W, VN, AGKE) - V 214/N 215 (VN) - ASP (Fu, W, VN, AGKE) - A 233/G 234/K 239/E 241 (AGKE) - AAL(Fu, W, VN, AGKE) - AIA (Fu, W, VN, AGKE)

Knockout strains – creating the experimental space First strains Utilizing ribose sugars Knock out strains W 3110 F- λ- IN (rrn. D-rrn. E)1 Problems * Wild type like strains showed unexpected recombination events Knockout strains * Wild type like strains showed difficulties to isolate plasmids Rpi. A-/B-: Collaboration Xyl. A-: Created own strain based on E. coli K 12: W 3110 Solution * Simple protocol by use of p. DK 43 expressing λ red recombinase and the p. CP 20 expressing FLP both a 43 o. C Ara. A-: based on E. coli K 12: W 3110 ongoing (Datsenko and Warren PNAS 2000) Materials and protocols were a kind gift from: Marinus G. Casteleijn Prof. R. Sterner 10 -11 February, Helsinki Dr. J. Claren KETJU meeting University of Regensburg

Selection – the use of the experimental space Libraries (16) - A (Fu, W, VN, AGKE) Selection Replacing known isomerase activity Cell - ASP (Fu, W, VN, AGKE) - AAL(Fu, W, VN, AGKE) Plate Loop 8 Neg. control - AIA (Fu, W, VN, AGKE) Neg. control Knockout strains RPIA-/B-: Collaboration Xyl. A-: Created own strain based on E. coli K 12: W 3110 Ara. A-: based on E. coli K 12: W 3110 ongoing Knock Out Marinus G. Casteleijn E. coli 10 -11 February, Helsinki Strain KETJU meeting Rondom gene from library Plasmid Pos. control L-Arabinose Isomerase Initial hits (4) for characterization. No Hits Two Hits However screening will be repeated with Ara. A- E. coli K 12: W 3110 strain. D-Xylose Isomerase Hits (2) for characterization Plate with selective media (i. e. One (1) carbon source) Positive controle gene Colony utilizing selective sugars

Biocatalysis at our Facilities The right Tools for the Right Methods. . . Tools New Methods High Throughput transformation * Hamilton pipetting station High Throughput optimization of protein expression Parallelization * Small scale cultivation technology (En. Base) * Parallel cloning library Miniaturization * Cultivations * Parallel cloning library Marinus G. Casteleijn 10 -11 February, Helsinki KETJU meeting 46 From Small Scale to Large Scale without further optimization High Throughput production of crystals for Crystallography ongoing
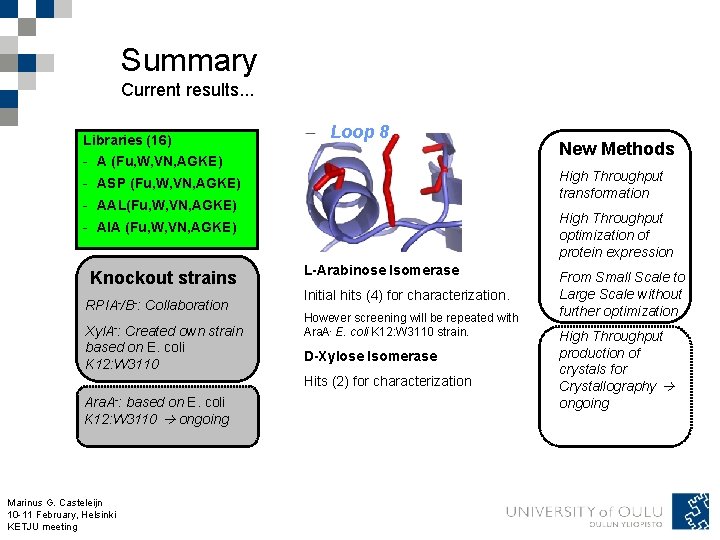
Summary Current results. . . Libraries (16) Loop 8 - A (Fu, W, VN, AGKE) High Throughput transformation - ASP (Fu, W, VN, AGKE) - AAL(Fu, W, VN, AGKE) High Throughput optimization of protein expression - AIA (Fu, W, VN, AGKE) Knockout strains RPIA-/B-: Collaboration Xyl. A-: Created own strain based on E. coli K 12: W 3110 L-Arabinose Isomerase Initial hits (4) for characterization. However screening will be repeated with Ara. A- E. coli K 12: W 3110 strain. D-Xylose Isomerase Hits (2) for characterization Ara. A-: based on E. coli K 12: W 3110 ongoing Marinus G. Casteleijn 10 -11 February, Helsinki KETJU meeting New Methods From Small Scale to Large Scale without further optimization High Throughput production of crystals for Crystallography ongoing
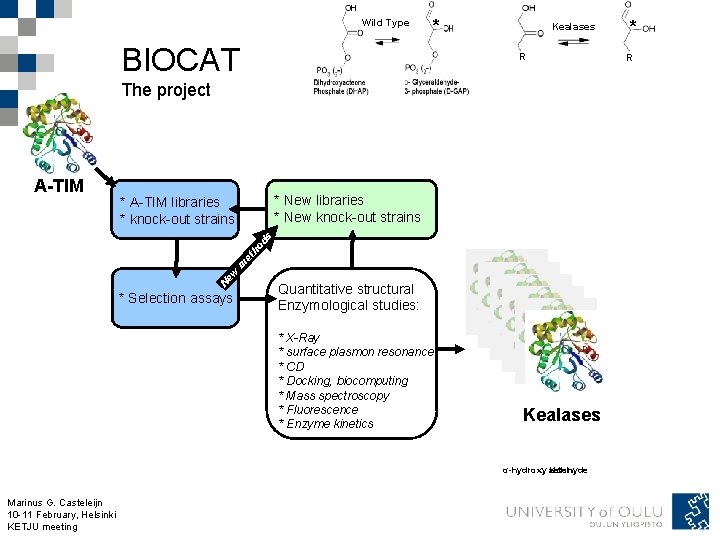
Wild Type BIOCAT * Kealases R The project * New libraries * New knock-out strains * A-TIM libraries * knock-out strains Ne w m et ho ds A-TIM * Selection assays Quantitative structural Enzymological studies: * X-Ray * surface plasmon resonance * CD * Docking, biocomputing * Mass spectroscopy * Fluorescence * Enzyme kinetics Kealases α-hydroxy aldehyde keton Marinus G. Casteleijn 10 -11 February, Helsinki KETJU meeting * R
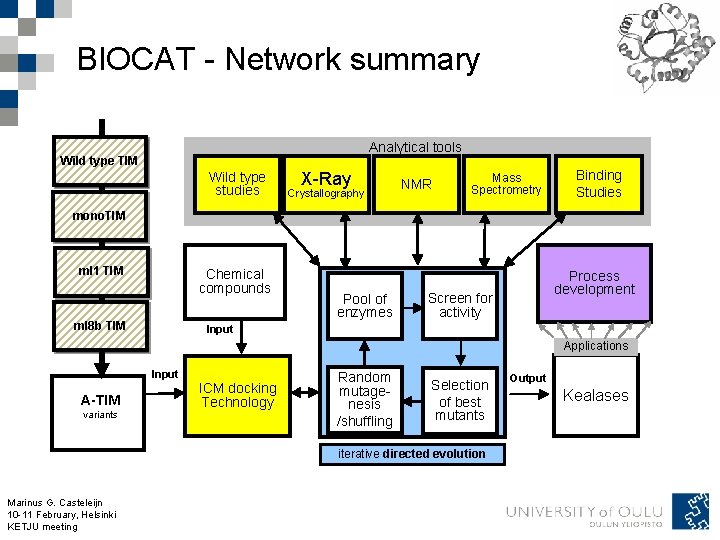
BIOCAT - Network summary Analytical tools Wild type TIM Wild type studies X-Ray Crystallography NMR Mass Spectrometry Binding Studies mono. TIM ml 1 TIM Chemical compounds ml 8 b TIM Pool of enzymes Process development Screen for activity Input Applications Input A-TIM variants ICM docking Technology Random mutagenesis /shuffling Selection of best mutants iterative directed evolution Marinus G. Casteleijn 10 -11 February, Helsinki KETJU meeting Output Kealases
- Slides: 13