Modeling molecular evolution Jodi Schwarz and Marc Smith






- Slides: 18

Modeling molecular evolution Jodi Schwarz and Marc Smith Vassar College Biol/CS 353 Bioinformatics

Biol / CS 353 Bioinformatics • Team taught Biol and Comp. Sci course • 7 students: – CS experience: 3 yes, 4 no – Bio experience: 5 yes, 2 no • Project-based course; no exams • Worked in Biol/CS pairs on projects • I 3 U near end of course; last project before independent research projects

Common approach for all projects • Biological question • Algorithm design – Step-by-step approach to complete a task or solve the problem • Implementation – The actual programming “script” that will carry out the steps of the algorithm • Evaluation of implementation and algorithm • Revision or augmentation

I 3 U: added an experimental component to our basic approach • Previous projects focused on pattern finding, mining whole genome data Goal of I 3 U: • Model a biological/evolutionary process • Test the model with empirical data • Perform computational experiments

Model molecular evolution • Step 1: model the effect of random vs targeted nucleotide substitutions on a protein sequence – What do we mean by random? – determine the similarity of the original protein sequence to the “evolved” sequence • Step 2: Assess the real nt diversity at positions 1, 2, 3 of codons in real homologs (HSP 70) – Construct alignment of homologs and determine nt diversity at each position • Evaluate the models using the empirical data

Learning goals • CS students: To apply their knowledge of data structures and algorithms to a biological domain • Biology students: To apply their knowledge of the biology to design algorithms • For the collaboration: – To become familiar with modeling a biological process: a simple model must be constructed and tested first – To test the model using empirical data

Assessment • Assignments – Alignment assignment – 2 Perl scripts • Model random vs targeted substitution pattern • Determine the codon nt diversity in HSP 70 genes – Output from the 2 Perl scripts • Raw output • Graphs summarizing data • Observation – Collaboration – Critical thinking

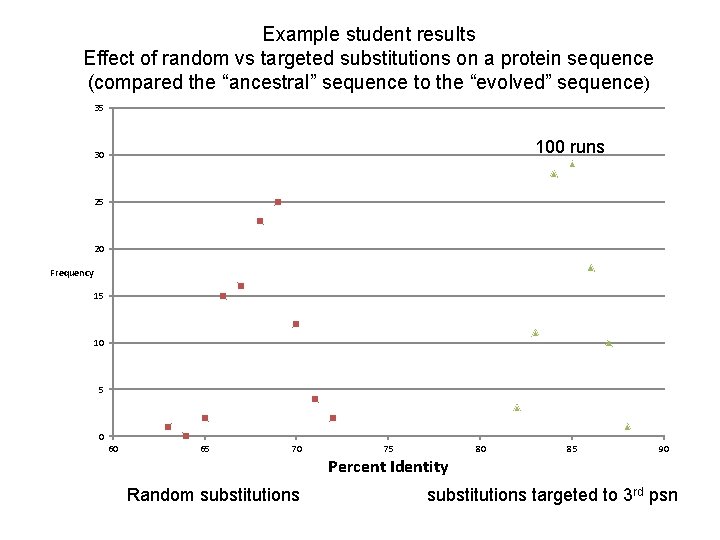
Example student results Effect of random vs targeted substitutions on a protein sequence (compared the “ancestral” sequence to the “evolved” sequence) 35 100 runs 30 25 20 Frequency 15 10 5 0 60 65 70 Random substitutions 75 Percent Identity 80 85 90 substitutions targeted to 3 rd psn

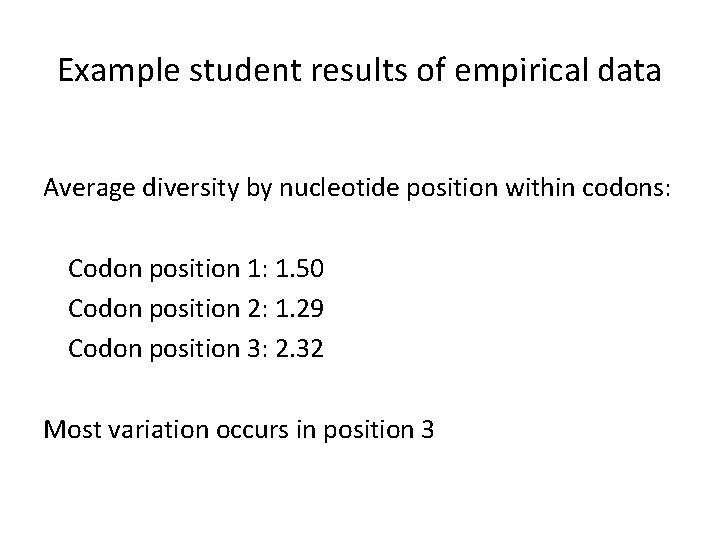
Example student results of empirical data Average diversity by nucleotide position within codons: Codon position 1: 1. 50 Codon position 2: 1. 29 Codon position 3: 2. 32 Most variation occurs in position 3

Collaboration across disciplines • How we tried to teach collaboration: – We defined the meaning of collaboration • • CS students do not need to become biologists and vice versa Each person contributes a different set of expertise Learning how to speak each other’s language Communication – We modeled it • Overt reliance on each other’s expertise • Spontaneous discussions – Giving students lots of experience collaborating: several shifts in pairs over the semester

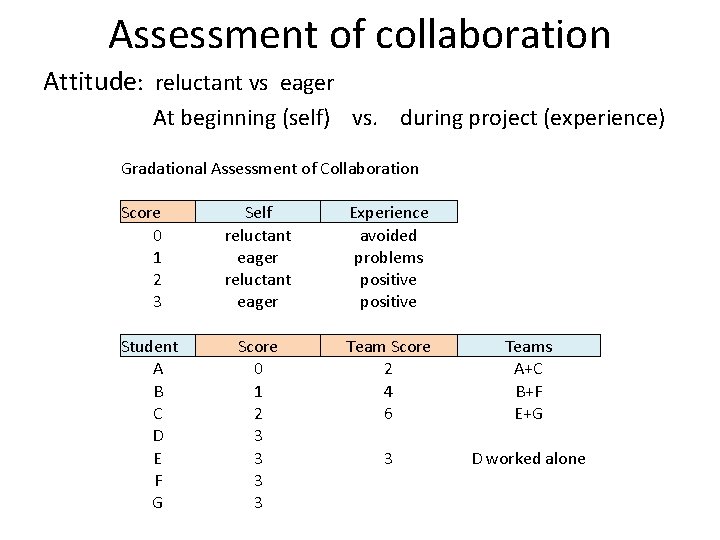
Assessment of collaboration Attitude: reluctant vs eager At beginning (self) vs. during project (experience) Gradational Assessment of Collaboration Score 0 1 2 3 Student A B C D E F G Self reluctant eager Experience avoided problems positive Score 0 1 2 3 3 Team Score 2 4 6 Teams A+C B+F E+G 3 D worked alone

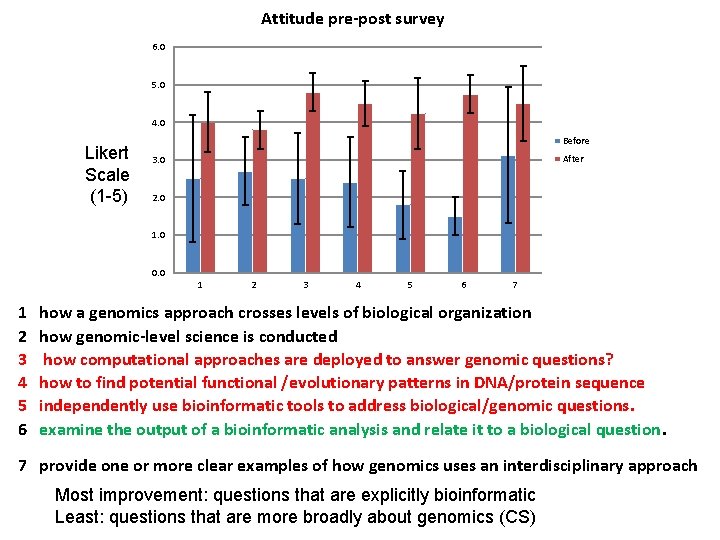
Attitude pre-post survey 6. 0 5. 0 4. 0 Likert Scale (1 -5) Before After 3. 0 2. 0 1. 0 0. 0 1 1 2 3 4 5 6 7 how a genomics approach crosses levels of biological organization how genomic-level science is conducted how computational approaches are deployed to answer genomic questions? how to find potential functional /evolutionary patterns in DNA/protein sequence independently use bioinformatic tools to address biological/genomic questions. examine the output of a bioinformatic analysis and relate it to a biological question. 7 provide one or more clear examples of how genomics uses an interdisciplinary approach Most improvement: questions that are explicitly bioinformatic Least: questions that are more broadly about genomics (CS)

What worked well • Overall approach was great: question, algorithm, implementation, analysis, iteration • Use of starter code allowed students to – Undertake much more sophisticated projects – see examples of more advanced algorithm/code • Encountering unanticipated results and problems – Gaps in alignments not in groups of 3 – Spontaneous discussions leading to AHA moments • Students enjoyed the modeling process – One student’s final project focused on modeling molecular evolution

What didn’t work as well • Some collaborations are not successful • Ran out of time: insufficient analysis and reflection • For the I 3 U: Assessment strategy not well developed – Can we retroactively extract more informative assessment?


Assessing biology knowledge • Algorithm development – Ability to help partner understand different mutation vs selection – Ability to recognize assumptions of model – Ability to use the empirical data to evaluate model

Assessing the CS • Variables – Abstraction: representing information as data – Types of data: predefined, atomic, aggregate – Scope: declaration, initialization, mutation • Algorithms – – Control flow: unconditional, repetition Input/Output and regex (pattern matching) Top-down design: subroutines To reuse or not to reuse (code)? • Incremental development / experimentation • Elegance: readability and maintainability

• Biological question – What pattern of nucleotide substitution occurs in protein-coding genes? • Algorithm – What does we know about mutation, nt/AA sequences? – Assumptions • Implementation – Instructors provided “starter code” – Students read and ran the code to see what it did – Pairs discussed how to add and refine it, and did so • Evaluation – Analyze the CS: Did it run and did it do the job we asked? – Analyze the biology: Did it accurately represent the biological process? • Testing the models against empirical evidence – Aligned HSP 70 genes and evaluated the pattern of substitution • Which model most closely matched the biology?