Model With Crossed and Nested Factors Moisture Content














- Slides: 21

Model With Crossed and Nested Factors Moisture Content of Tree Branch Cuts Mc. Dermott, J. J. (1941). “The Effect of the Method of Cutting on the Moisture Content of Samples from Tree Branches”, American Journal of Botany, Vol. 28, #6, pp. 506 -508.

Model Description • Response: Moisture Content (10 x% of Dry weight) • Factors: – Species (5 Replicate Branches per Species) • • Loblolly Pine Shortleaf Pine Yellow Poplar Red Gum – Branch Locations • Central • Distal • Proximal – Transpiration Types • Rapid (Hot, Dry, Sunny Day) • Slow (Cool, Moist, Cloudy Day)

Statistical Model

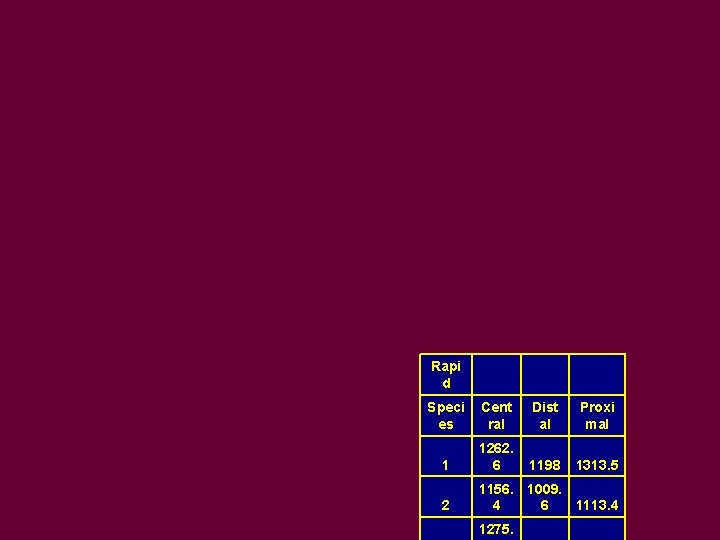
Rapi d Speci es Cent ral Dist al Proxi mal 1 1262. 6 1198 1313. 5 2 1156. 1009. 4 6 1113. 4 1275.

Slow Species Central Distal Proximal 1 1196 1257. 6 1262. 6 2 1036. 2 1178 1156. 4 3 1122. 6 1180. 2 1275. 8 4 1124. 6 1142. 8 1258. 6

Analysis of Variance (Main/Nested Effects)

Analysis of Variance (Interaction Terms)

Expected Mean Squares (Main/Nested Effects)

Expected Mean Squares (Interaction Terms)

Testing for Interaction Effects

Testing for Main/Nested Effects

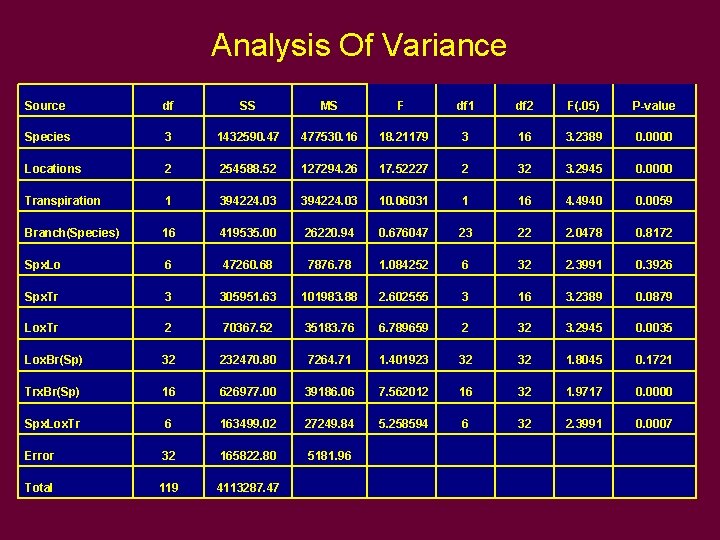
Analysis Of Variance Source df SS MS F df 1 df 2 F(. 05) P-value Species 3 1432590. 47 477530. 16 18. 21179 3 16 3. 2389 0. 0000 Locations 2 254588. 52 127294. 26 17. 52227 2 32 3. 2945 0. 0000 Transpiration 1 394224. 03 10. 06031 1 16 4. 4940 0. 0059 Branch(Species) 16 419535. 00 26220. 94 0. 676047 23 22 2. 0478 0. 8172 Spx. Lo 6 47260. 68 7876. 78 1. 084252 6 32 2. 3991 0. 3926 Spx. Tr 3 305951. 63 101983. 88 2. 602555 3 16 3. 2389 0. 0879 Lox. Tr 2 70367. 52 35183. 76 6. 789659 2 32 3. 2945 0. 0035 Lox. Br(Sp) 32 232470. 80 7264. 71 1. 401923 32 32 1. 8045 0. 1721 Trx. Br(Sp) 16 626977. 00 39186. 06 7. 562012 16 32 1. 9717 0. 0000 Spx. Lox. Tr 6 163499. 02 27249. 84 5. 258594 6 32 2. 3991 0. 0007 Error 32 165822. 80 5181. 96 Total 119 4113287. 47

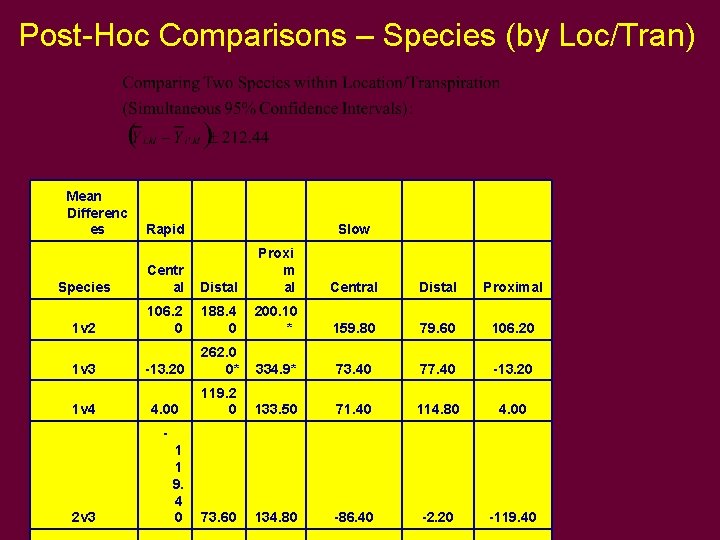
Post-Hoc Comparisons – Species (by Loc/Tran)

Post-Hoc Comparisons – Species (by Loc/Tran)

Post-Hoc Comparisons – Species (by Loc/Tran) Mean Differenc es Slow Species Centr al Distal Proxi m al Central Distal Proximal 1 v 2 106. 2 0 188. 4 0 200. 10 * 159. 80 79. 60 106. 20 -13. 20 262. 0 0* 334. 9* 73. 40 77. 40 -13. 20 4. 00 119. 2 0 133. 50 71. 40 114. 80 4. 00 73. 60 134. 80 -86. 40 -2. 20 -119. 40 1 v 3 1 v 4 Rapid - 2 v 3 1 1 9. 4 0

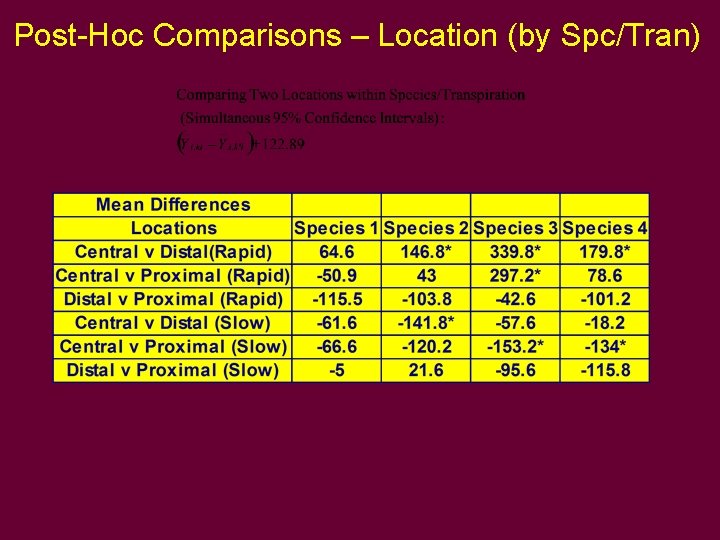
Post-Hoc Comparisons – Locations (by Spe/Tran)

Post-Hoc Comparisons – Locations (by Spe/Tran)

Post-Hoc Comparisons – Location (by Spc/Tran)

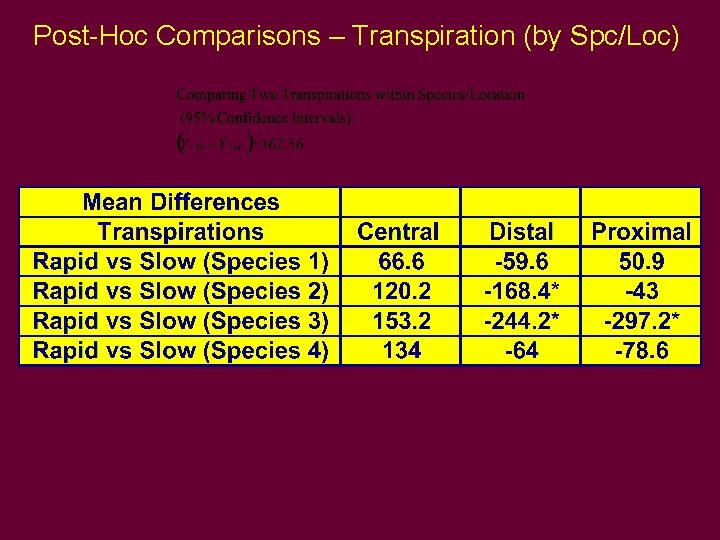
Post-Hoc Comparisons – Transpirations (by Spe/Loc)

Post-Hoc Comparisons – Transpirations (by Spe/Loc)

Post-Hoc Comparisons – Transpiration (by Spc/Loc)