MODEL BASED DEEP LEARNING IN FREE BREATHING UNGATED
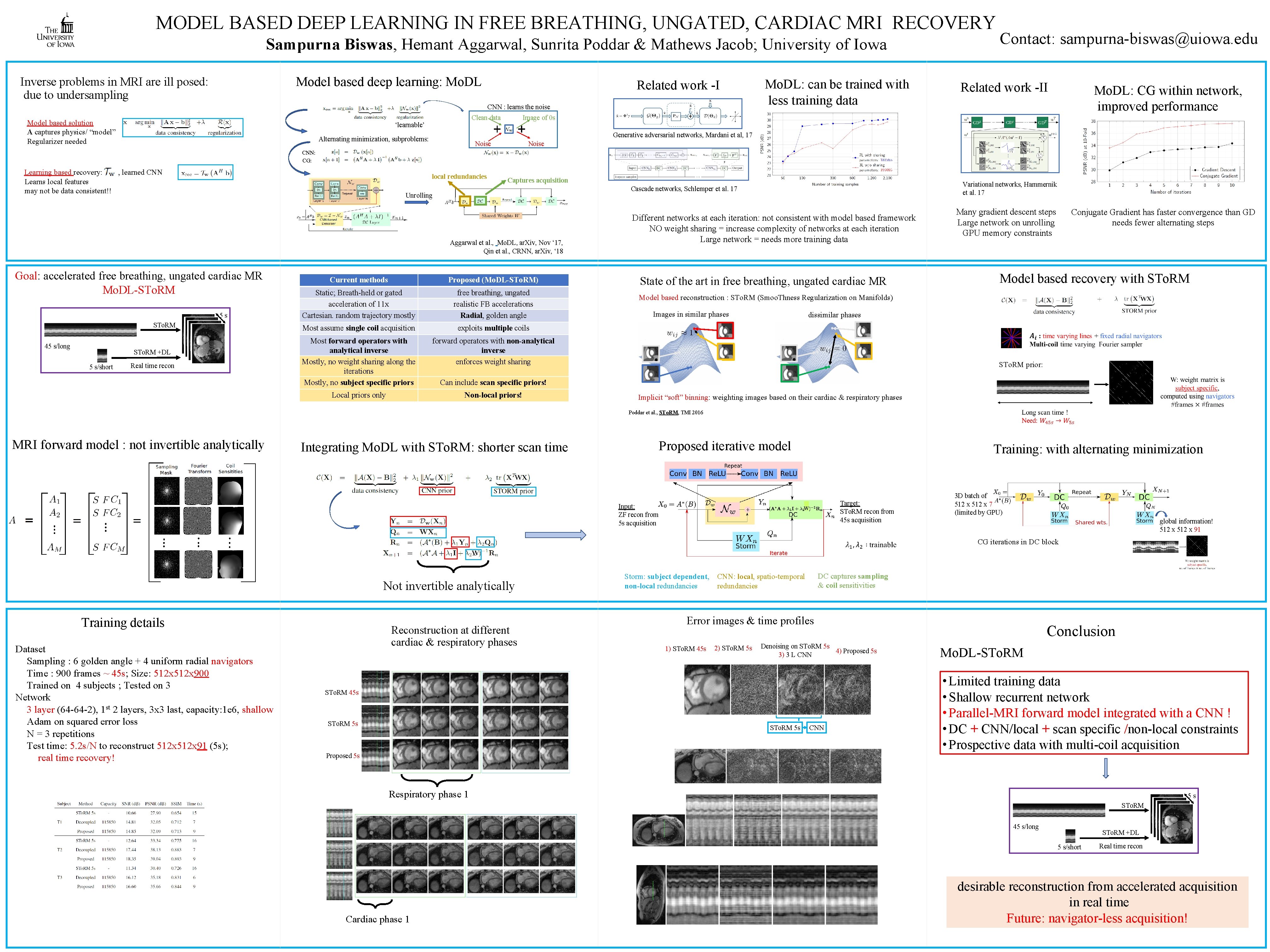
MODEL BASED DEEP LEARNING IN FREE BREATHING, UNGATED, CARDIAC MRI RECOVERY Sampurna Biswas, Hemant Aggarwal, Sunrita Poddar & Mathews Jacob; University of Iowa Model based deep learning: Mo. DL Inverse problems in MRI are ill posed: due to undersampling Related work -I CNN : learns the noise Model based solution A captures physics/ “model” Regularizer needed Clean data ‘learnable’ Mo. DL: can be trained with less training data Contact: sampurna-biswas@uiowa. edu Related work -II Mo. DL: CG within network, improved performance Image of 0 s Generative adversarial networks, Mardani et al, 17 Alternating minimization, subproblems: Noise CNN: CG: Learning based recovery: , learned CNN Learns local features may not be data consistent!! local redundancies Captures acquisition Unrolling Aggarwal et al. , Mo. DL, ar. Xiv, Nov ‘ 17, Qin et al. , CRNN, ar. Xiv, ‘ 18 Goal: accelerated free breathing, ungated cardiac MR Mo. DL-STo. RM 5 s STo. RM 45 s/long STo. RM +DL 5 s/short Real time recon Variational networks, Hammernik et al. 17 Cascade networks, Schlemper et al. 17 Current methods Proposed (Mo. DL-STo. RM) Static; Breath-held or gated acceleration of 11 x Cartesian. random trajectory mostly free breathing, ungated realistic FB accelerations Radial, golden angle Most assume single coil acquisition exploits multiple coils Most forward operators with analytical inverse Mostly, no weight sharing along the iterations Mostly, no subject specific priors forward operators with non-analytical inverse enforces weight sharing Local priors only Non-local priors! Different networks at each iteration: not consistent with model based framework NO weight sharing = increase complexity of networks at each iteration Large network = needs more training data State of the art in free breathing, ungated cardiac MR Many gradient descent steps Large network on unrolling GPU memory constraints Conjugate Gradient has faster convergence than GD needs fewer alternating steps Model based recovery with STo. RM Model based reconstruction : STo. RM (Smoo. Thness Regularization on Manifolds) Images in similar phases dissimilar phases STo. RM prior: Can include scan specific priors! Implicit “soft” binning: weighting images based on their cardiac & respiratory phases Poddar et al. , STo. RM, TMI 2016 MRI forward model : not invertible analytically Proposed iterative model Integrating Mo. DL with STo. RM: shorter scan time Input: ZF recon from 5 s acquisition Training: with alternating minimization Target: STo. RM recon from 45 s acquisition W 3 D batch of 512 x 7 (limited by GPU) global information! 512 x 91 CG iterations in DC block Not invertible analytically Training details Dataset Sampling : 6 golden angle + 4 uniform radial navigators Time : 900 frames ~ 45 s; Size: 512 x 900 Trained on 4 subjects ; Tested on 3 Network 3 layer (64 -64 -2), 1 st 2 layers, 3 x 3 last, capacity: 1 e 6, shallow Adam on squared error loss N = 3 repetitions Test time: 5. 2 s/N to reconstruct 512 x 91 (5 s); real time recovery! Reconstruction at different cardiac & respiratory phases Storm: subject dependent, non-local redundancies DC captures sampling & coil sensitivities CNN: local, spatio-temporal redundancies Error images & time profiles 1) STo. RM 45 s 2) STo. RM 5 s Denoising on STo. RM 5 s 4) Proposed 5 s 3) 3 L CNN STo. RM 45 s STo. RM 5 s CNN Conclusion Mo. DL-STo. RM • Limited training data • Shallow recurrent network • Parallel-MRI forward model integrated with a CNN ! • DC + CNN/local + scan specific /non-local constraints • Prospective data with multi-coil acquisition Proposed 5 s Respiratory phase 1 5 s STo. RM 45 s/long STo. RM +DL 5 s/short Cardiac phase 1 Real time recon desirable reconstruction from accelerated acquisition in real time Future: navigator-less acquisition!
- Slides: 1