MNTP Trainee Georgina Vinyes Junque Chi Hun Kim





- Slides: 18

MNTP Trainee: Georgina Vinyes Junque, Chi Hun Kim Prof. James T. Becker Cyrus Raji, Leonid Teverovskiy, and Robert Tamburo Anatomical MRI module

Voxel-Based Morphometry (VBM) Structural differences based on Voxel-wise comparision Advantages Automated, Un-biased, Whole brain analysis compared to Manual ROI tracing Well established and Widely used over the past decade Results are biologically plausible and replicable We know the LIMITATIONS

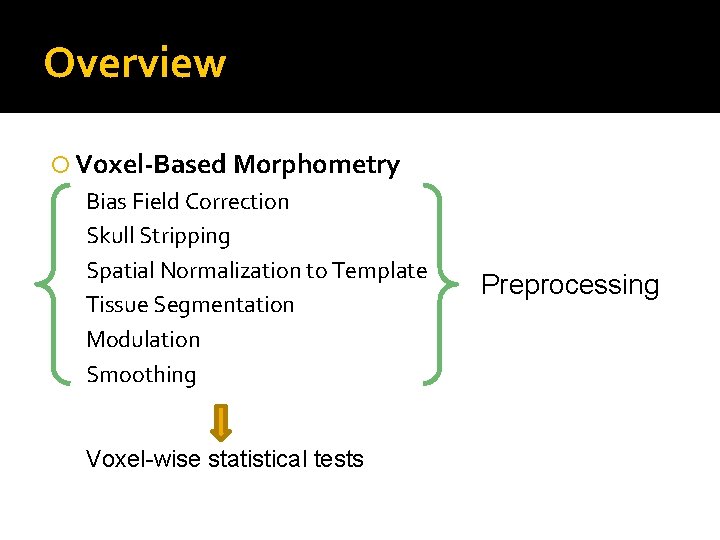
Overview Voxel-Based Morphometry Bias Field Correction Skull Stripping Spatial Normalization to Template Tissue Segmentation Modulation Smoothing Voxel-wise statistical tests Preprocessing

Methods MRI sequence T 1 (MPRAGE) Multicenter AIDS Cohort Study (MACS) 53 males Age: 50. 2 +- 4. 4 3 T Siemens Trio. Tim Slices: 160; thickness 1. 2 mm Voxel size: 1 x 1. 2 mm TE: 2. 98; TR: 2300 Software SPM 2 & SPM 5 (Wellcome Trust Centre for Neuroimaging) VBM 2 toolbox (Gaser et al, http: //dbm. neuro. uni-jena. de/) N 3 algorithm Brain Extraction Tool in FSL Watershed algorithm in Free. Surfer Subjects Statistical Analysis Gray matter Volume differences in Drug users vs. Non-Drug users

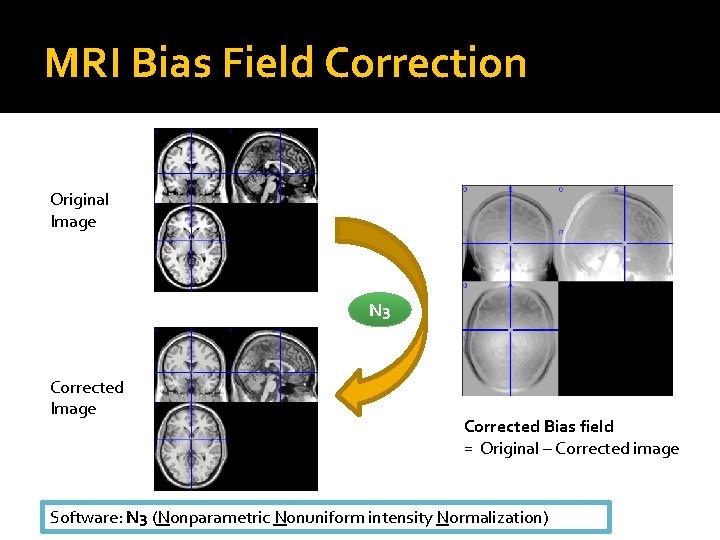
MRI Bias Field Correction Original Image N 3 Corrected Image Corrected Bias field = Original – Corrected image Software: N 3 (Nonparametric Nonuniform intensity Normalization)

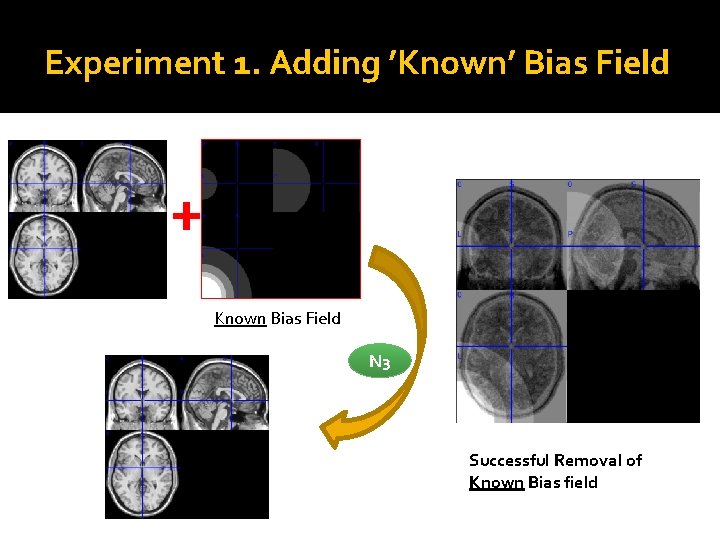
Experiment 1. Adding ’Known’ Bias Field + Known Bias Field N 3 Successful Removal of Known Bias field

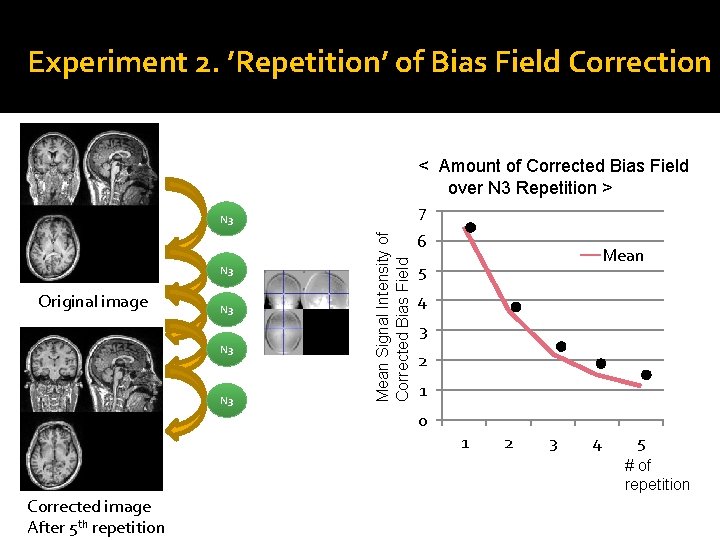
Experiment 2. ’Repetition’ of Bias Field Correction < Amount of Corrected Bias Field over N 3 Repetition > 7 N 3 Original image N 3 N 3 Mean Signal Intensity of Corrected Bias Field N 3 6 Mean 5 4 3 2 1 0 1 2 3 4 5 # of repetition Corrected image After 5 th repetition

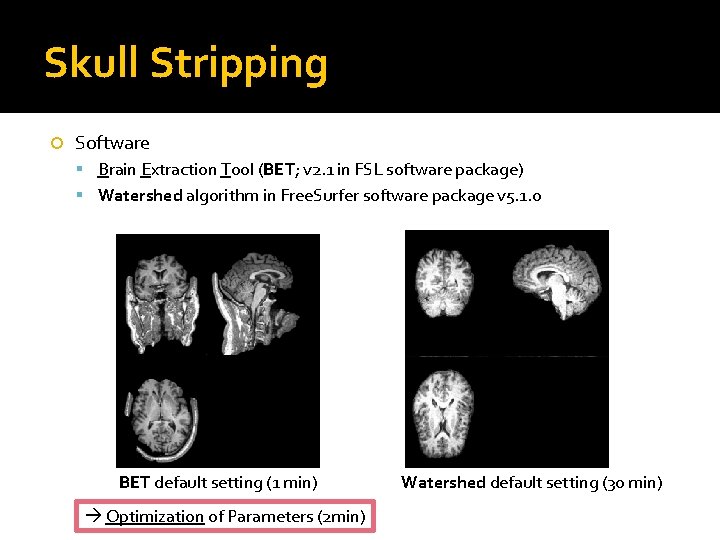
Skull Stripping Software Brain Extraction Tool (BET; v 2. 1 in FSL software package) Watershed algorithm in Free. Surfer software package v 5. 1. 0 BET default setting (1 min) Optimization of Parameters (2 min) Watershed default setting (30 min)

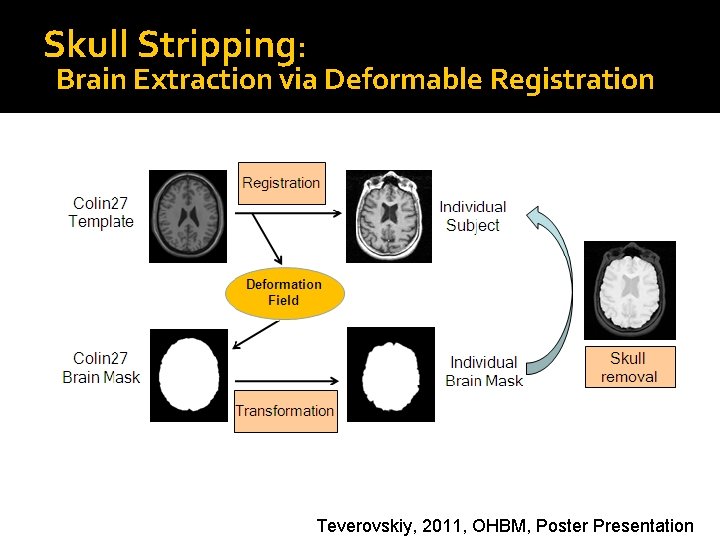
Skull Stripping: Brain Extraction via Deformable Registration Teverovskiy, 2011, OHBM, Poster Presentation

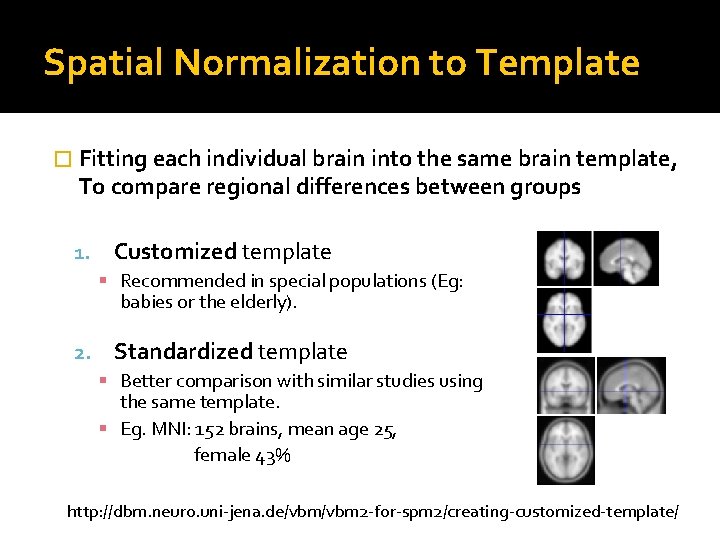
Spatial Normalization to Template � Fitting each individual brain into the same brain template, To compare regional differences between groups 1. Customized template Recommended in special populations (Eg: babies or the elderly). 2. Standardized template Better comparison with similar studies using the same template. Eg. MNI: 152 brains, mean age 25, female 43% http: //dbm. neuro. uni-jena. de/vbm 2 -for-spm 2/creating-customized-template/

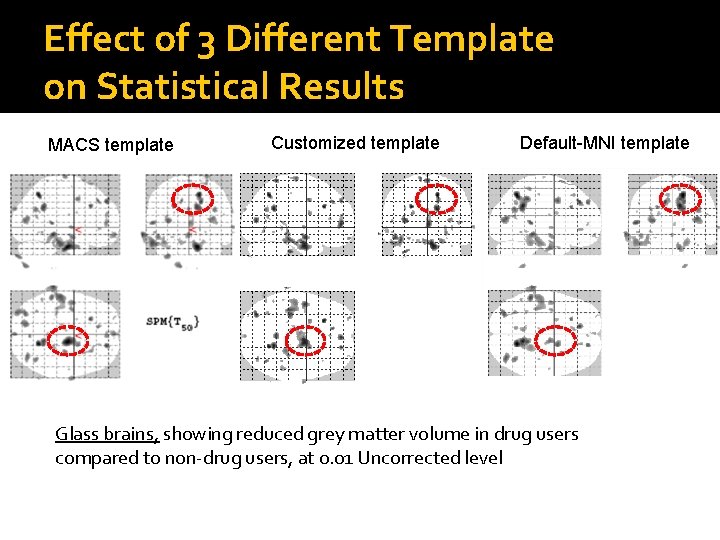
Effect of 3 Different Template on Statistical Results MACS template Customized template Default-MNI template Glass brains, showing reduced grey matter volume in drug users compared to non-drug users, at 0. 01 Uncorrected level

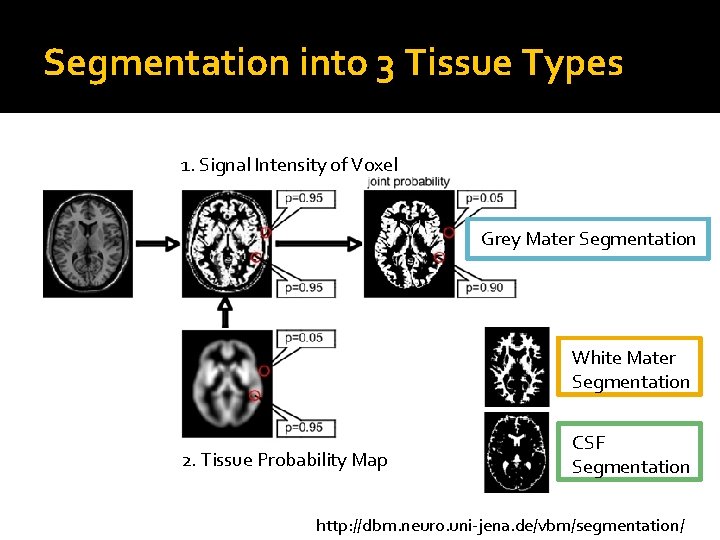
Segmentation into 3 Tissue Types 1. Signal Intensity of Voxel Grey Mater Segmentation White Mater Segmentation 2. Tissue Probability Map CSF Segmentation http: //dbm. neuro. uni-jena. de/vbm/segmentation/

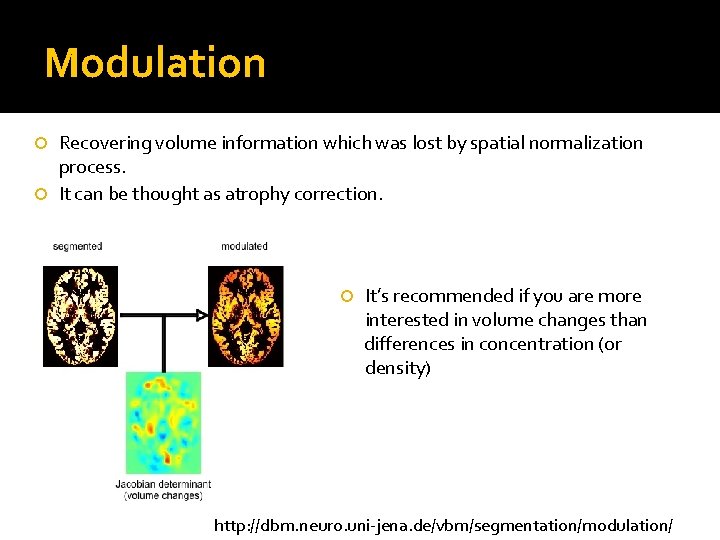
Modulation Recovering volume information which was lost by spatial normalization process. It can be thought as atrophy correction. It’s recommended if you are more interested in volume changes than differences in concentration (or density) http: //dbm. neuro. uni-jena. de/vbm/segmentation/modulation/

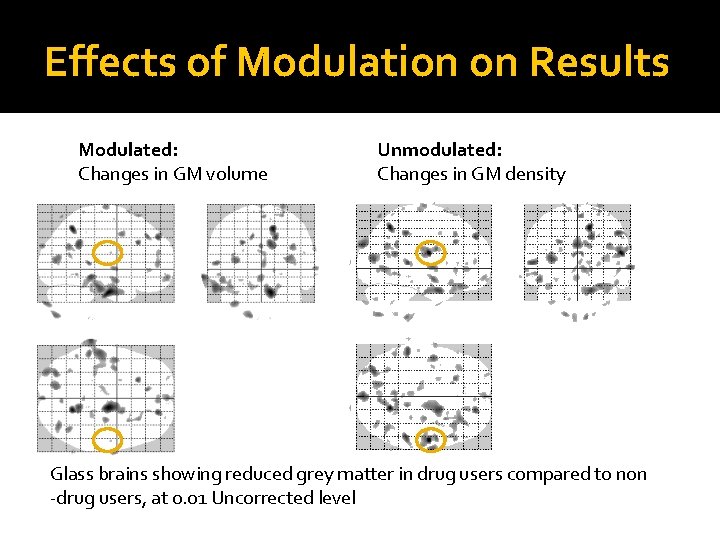
Effects of Modulation on Results Modulated: Changes in GM volume Unmodulated: Changes in GM density Glass brains showing reduced grey matter in drug users compared to non -drug users, at 0. 01 Uncorrected level

Smoothing Intensity of every voxel is replaced by the weighted average of the surrounding voxels. Larger kernel size, more surrounding voxels Make distribution closely to Gaussian field model Increase the sensitivity of tests by reducing the variance across subjects Reduce the effect of misregistration

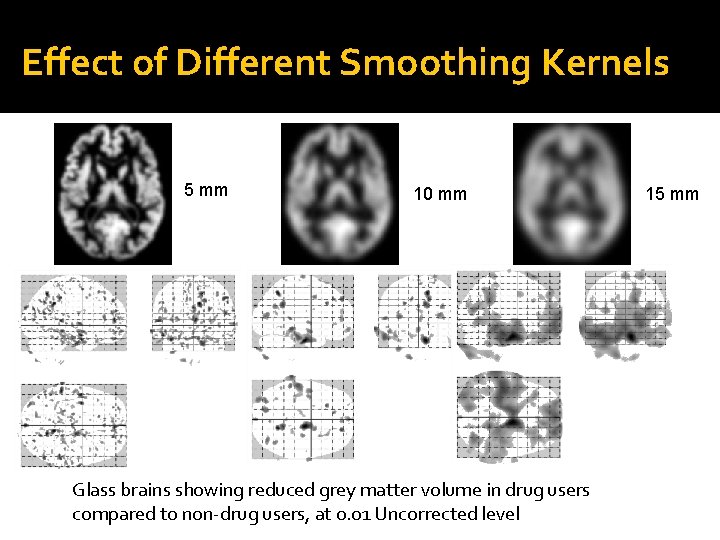
Effect of Different Smoothing Kernels 5 mm 10 mm Glass brains showing reduced grey matter volume in drug users compared to non-drug users, at 0. 01 Uncorrected level 15 mm

Conclusion There’s a lot of options in processing that can affect data and results. We have to undertand what we are doing in every step to better adjust options to our sample study. Since these techniques have several pitfalls, we have to carefully interpret published results.

Thank You Prof. James T. Becker TA: Cyrus Raji, Leonid Teverovskiy, Robert Tamburo Prof. Seong-Gi Kim & Prof. Bill Eddy Tomika Cohen, Rebecca Clark Fellow MNTPers!