Mitosis and the Cell Cycle Interphase and the






- Slides: 59

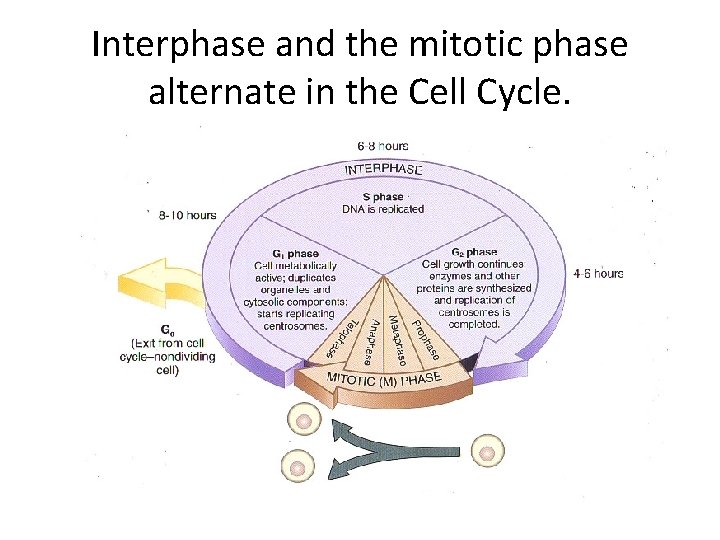
Mitosis and the Cell Cycle


Interphase and the mitotic phase alternate in the Cell Cycle.



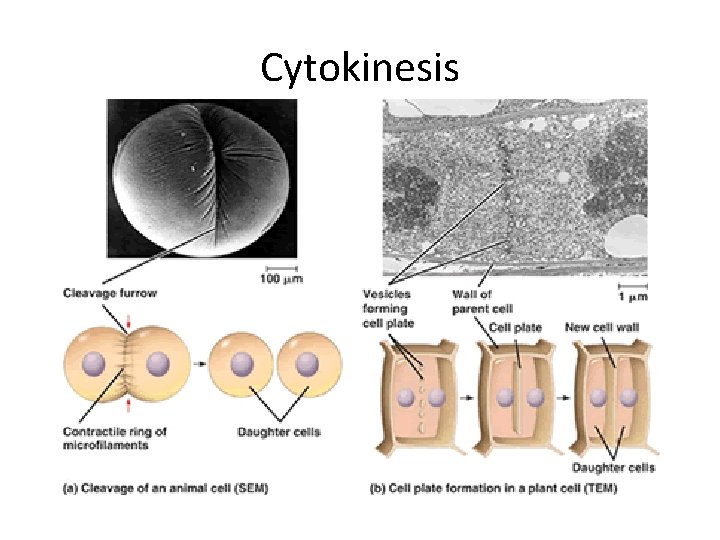
Cytokinesis






Meiosis

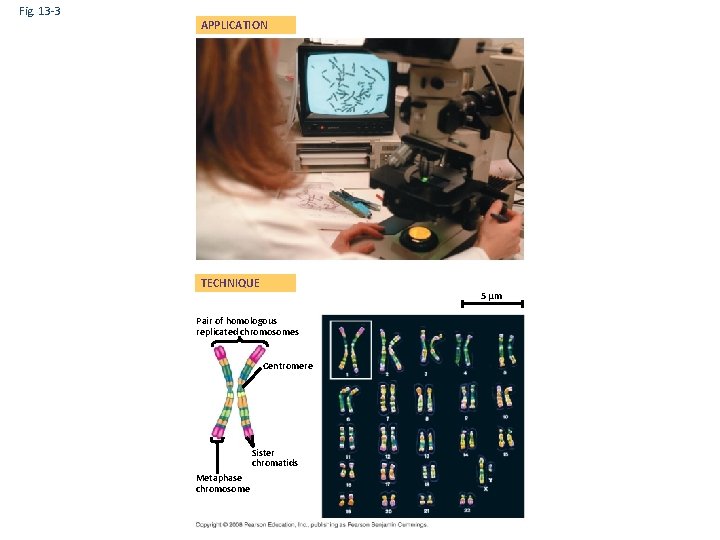
Fig. 13 -3 APPLICATION TECHNIQUE 5 µm Pair of homologous replicated chromosomes Centromere Sister chromatids Metaphase chromosome

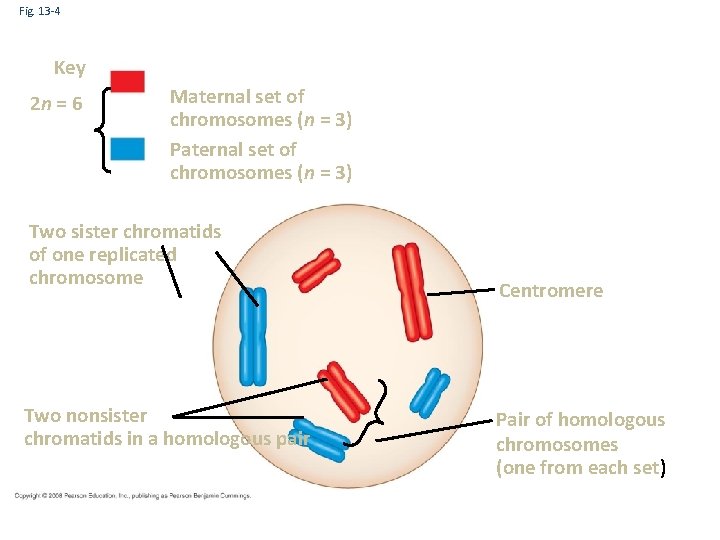
Fig. 13 -4 Key 2 n = 6 Maternal set of chromosomes (n = 3) Paternal set of chromosomes (n = 3) Two sister chromatids of one replicated chromosome Two nonsister chromatids in a homologous pair Centromere Pair of homologous chromosomes (one from each set)

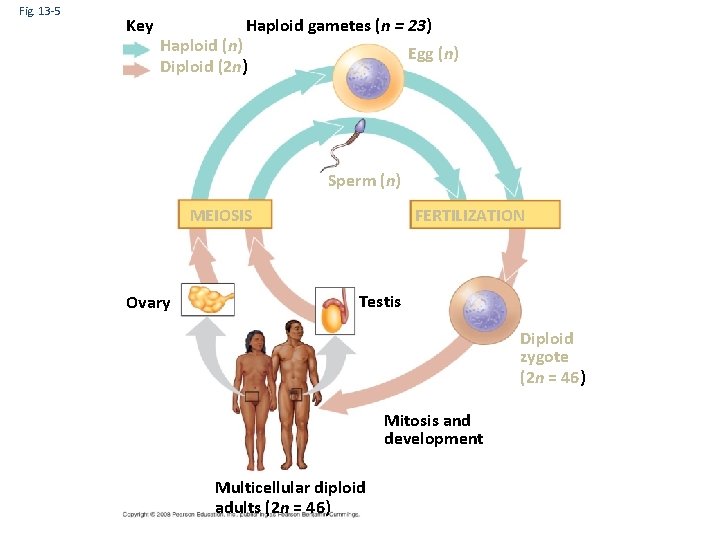
Fig. 13 -5 Key Haploid gametes (n = 23) Haploid (n) Diploid (2 n) Egg (n) Sperm (n) MEIOSIS Ovary FERTILIZATION Testis Diploid zygote (2 n = 46) Mitosis and development Multicellular diploid adults (2 n = 46)

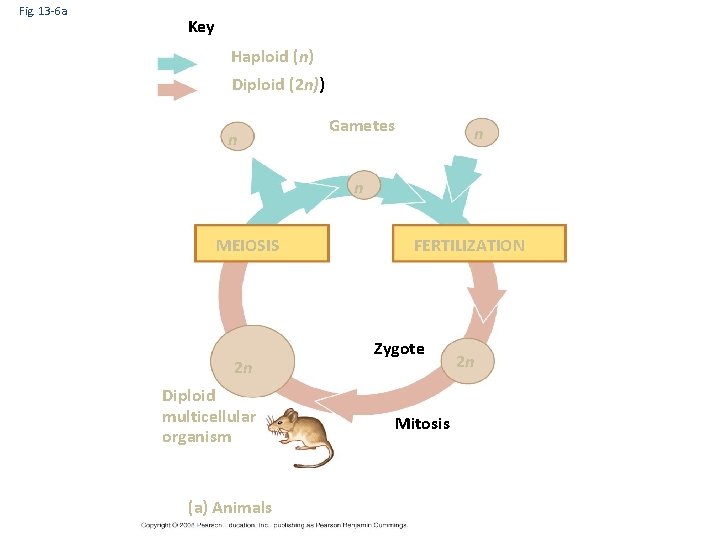
Fig. 13 -6 a Key Haploid (n) Diploid (2 n)) n Gametes n n MEIOSIS 2 n Diploid multicellular organism (a) Animals FERTILIZATION Zygote Mitosis 2 n

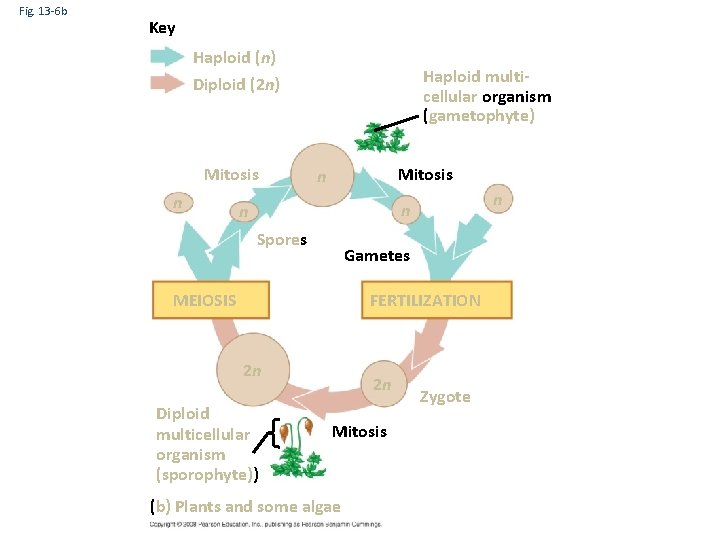
Fig. 13 -6 b Key Haploid (n) Diploid (2 n) Mitosis n Haploid multicellular organism (gametophyte) Mitosis n n Spores Gametes MEIOSIS FERTILIZATION 2 n Diploid multicellular organism (sporophyte)) 2 n Mitosis (b) Plants and some algae Zygote

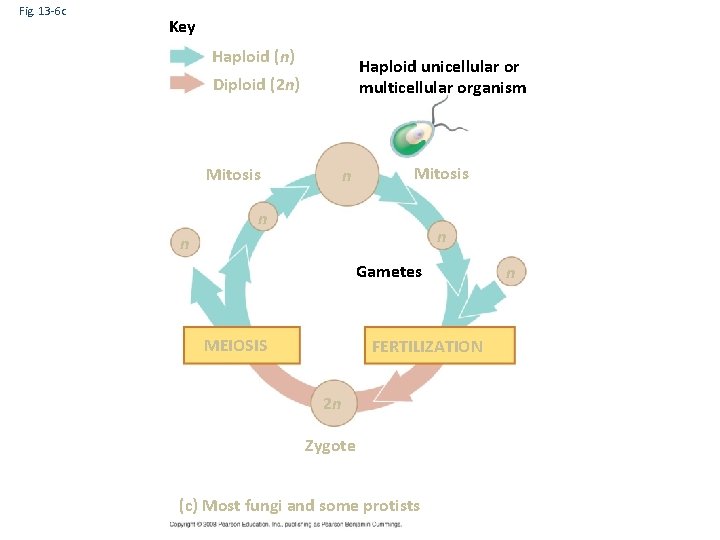
Fig. 13 -6 c Key Haploid (n) Haploid unicellular or multicellular organism Diploid (2 n) Mitosis n n n Gametes MEIOSIS FERTILIZATION 2 n Zygote (c) Most fungi and some protists n

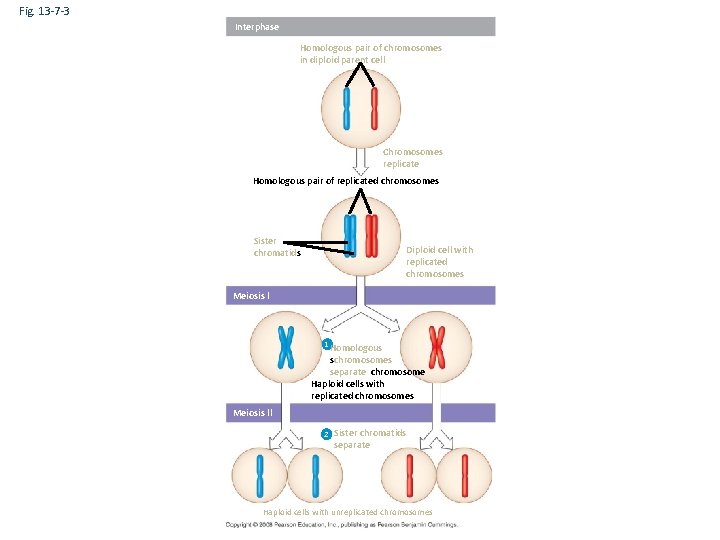
Fig. 13 -7 -3 Interphase Homologous pair of chromosomes in diploid parent cell Chromosomes replicate Homologous pair of replicated chromosomes Sister chromatids Diploid cell with replicated chromosomes Meiosis I 1 Homologous schromosomes separate chromosome Haploid cells with replicated chromosomes Meiosis II 2 Sister chromatids separate Haploid cells with unreplicated chromosomes

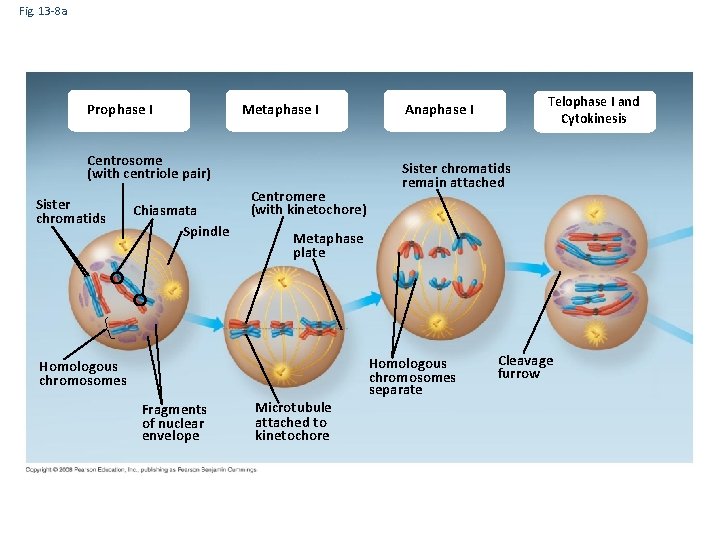
Fig. 13 -8 a Prophase I Metaphase I Centrosome (with centriole pair) Sister chromatids Chiasmata Spindle Centromere (with kinetochore) Sister chromatids remain attached Metaphase plate Homologous chromosomes Fragments of nuclear envelope Telophase I and Cytokinesis Anaphase I Microtubule attached to kinetochore Homologous chromosomes separate Cleavage furrow

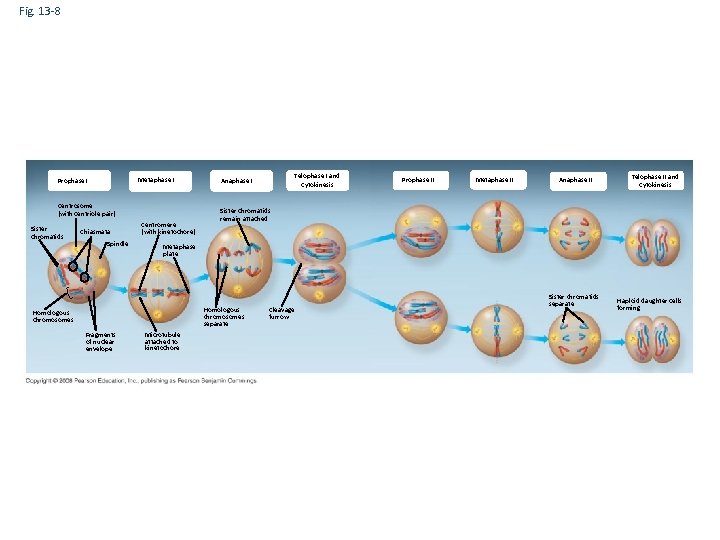
Fig. 13 -8 Metaphase I Prophase I Centrosome (with centriole pair) Sister chromatids Chiasmata Spindle Centromere (with kinetochore) Prophase II Metaphase II Anaphase II Telophase II and Cytokinesis Sister chromatids remain attached Metaphase plate Homologous chromosomes separate Homologous chromosomes Fragments of nuclear envelope Telophase I and Cytokinesis Anaphase I Microtubule attached to kinetochore Cleavage furrow Sister chromatids separate Haploid daughter cells forming

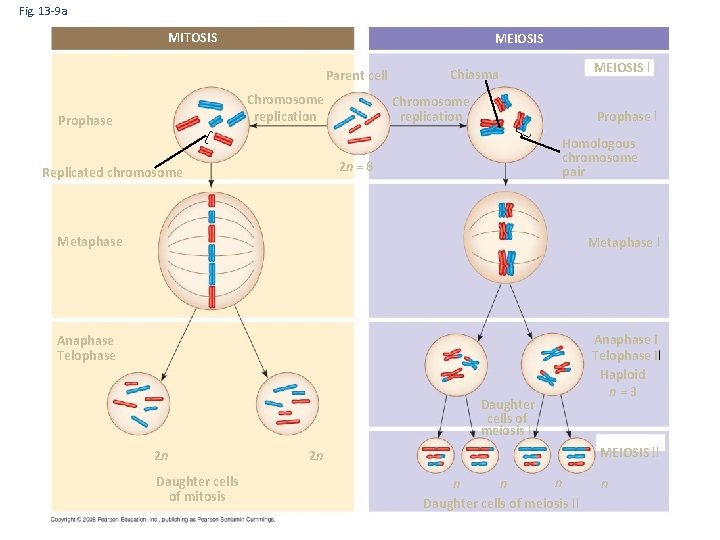
Fig. 13 -9 a MITOSIS MEIOSIS Parent cell Chromosome replication Prophase I Homologous chromosome pair 2 n = 6 Replicated chromosome MEIOSIS I Chiasma Metaphase I Anaphase Telophase Anaphase I Telophase II Haploid n=3 Daughter cells of meiosis I 2 n Daughter cells of mitosis MEIOSIS II 2 n n Daughter cells of meiosis II n

Fig. 13 -9 b SUMMARY Property Mitosis Meiosis DNA replication Occurs during interphase before mitosis begins Occurs during interphase before meiosis I begins Number of divisions One, including prophase, metaphase, and telophase Two, each including prophase, metaphase, and telophase Synapsis of homologous chromosomes Does not occur Occurs during prophase I along with crossing over between nonsister chromatids; resulting chiasmata hold pairs together due to sister chromatid cohesion Number of daughter cells and genetic composition Two, each diploid (2 n) and genetically identical to the parent cell Four, each haploid (n), containing half as many chromosomes as the parent cell; genetically different from the parent cell and from each other Role in the animal body Enables multicellular adult to arise from zygote; produces cells for growth, repair, and, in some species, asexual reproduction Produces gametes; reduces number of chromosomes by half and introduces genetic variability among the gametes

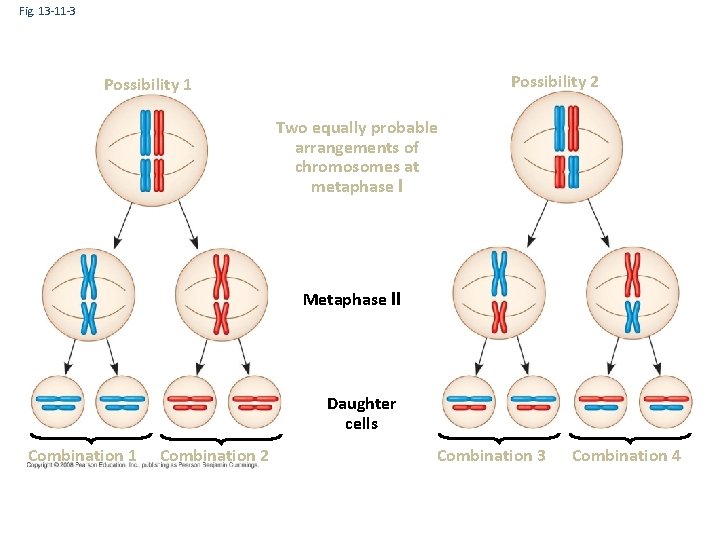
Fig. 13 -11 -3 Possibility 2 Possibility 1 Two equally probable arrangements of chromosomes at metaphase I Metaphase II Daughter cells Combination 1 Combination 2 Combination 3 Combination 4

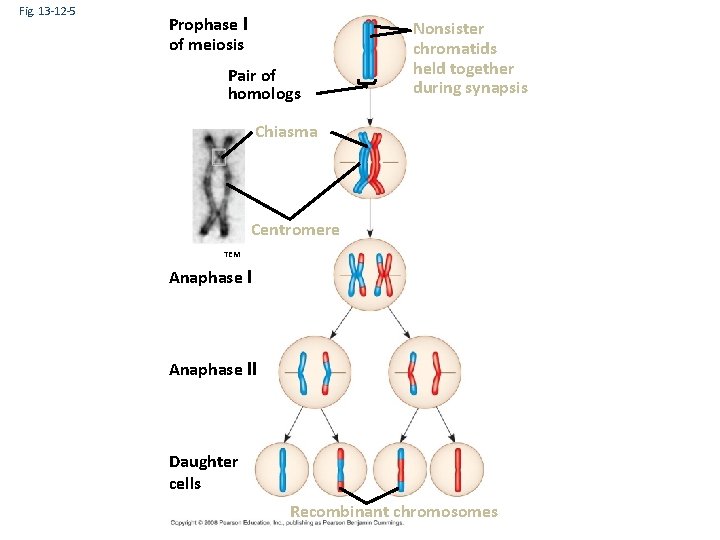
Fig. 13 -12 -5 Prophase I of meiosis Pair of homologs Nonsister chromatids held together during synapsis Chiasma Centromere TEM Anaphase II Daughter cells Recombinant chromosomes

Mendelian Genetics

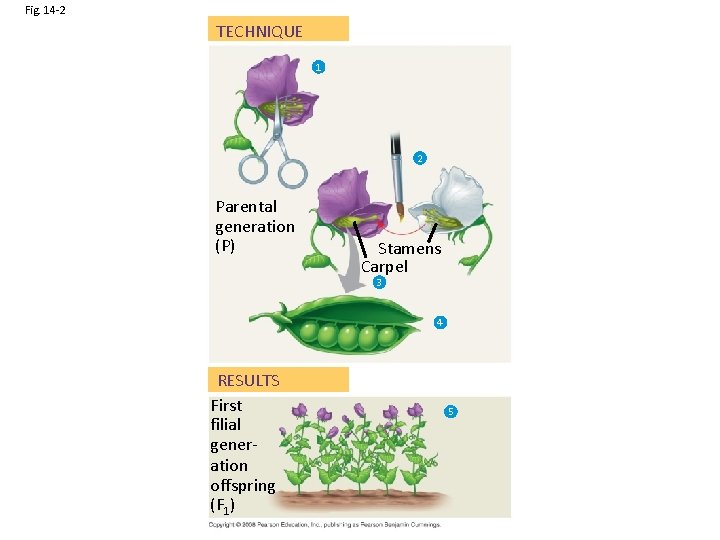
Fig. 14 -2 TECHNIQUE 1 2 Parental generation (P) Stamens Carpel 3 4 RESULTS First filial generation offspring (F 1) 5

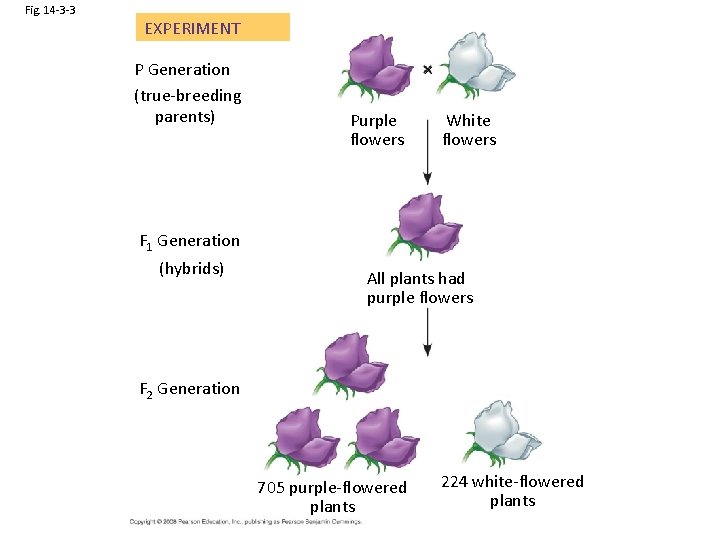
Fig. 14 -3 -3 EXPERIMENT P Generation (true-breeding parents) Purple flowers White flowers F 1 Generation (hybrids) All plants had purple flowers F 2 Generation 705 purple-flowered plants 224 white-flowered plants

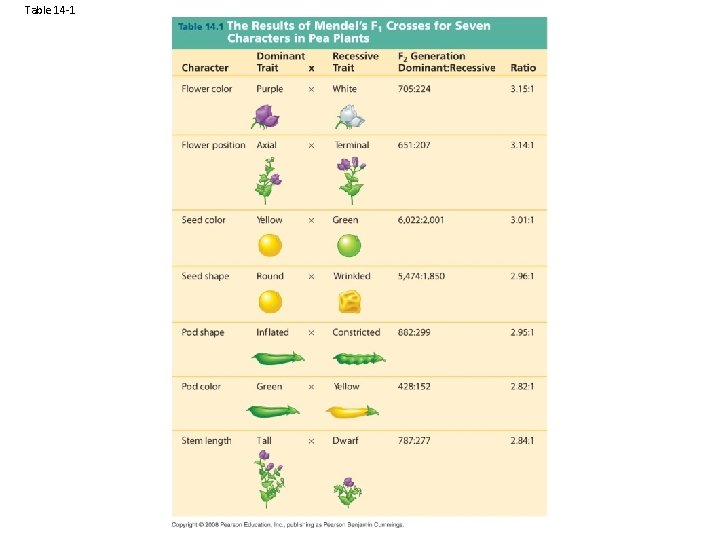
Table 14 -1

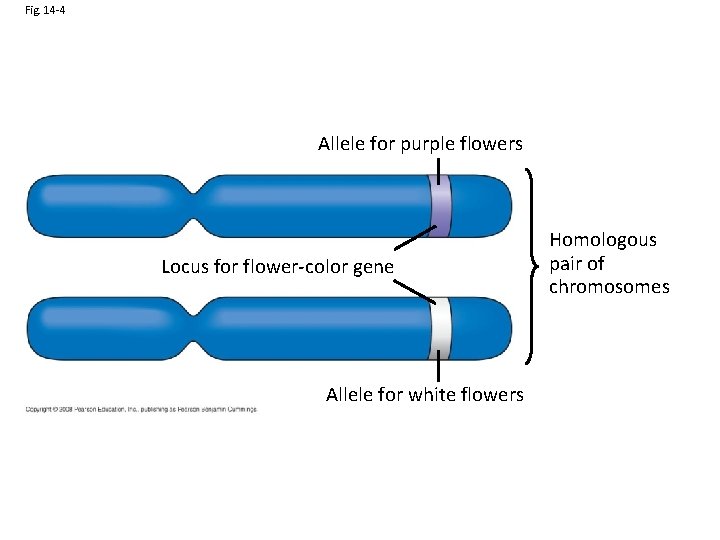
Fig. 14 -4 Allele for purple flowers Locus for flower-color gene Allele for white flowers Homologous pair of chromosomes

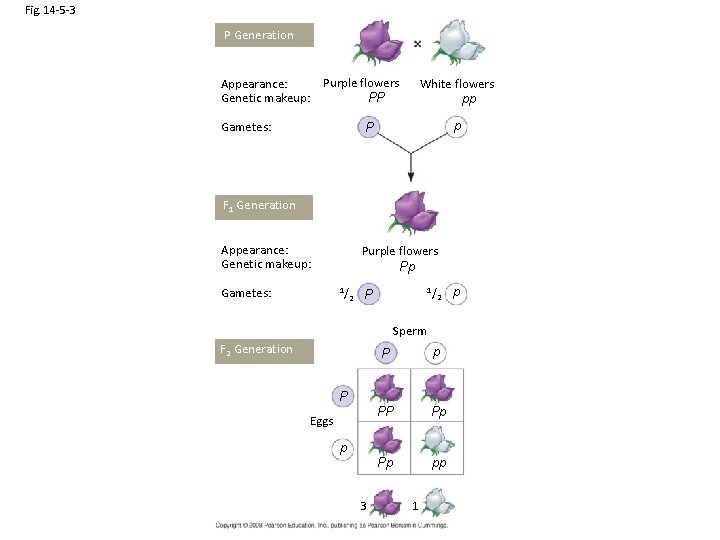
Fig. 14 -5 -3 P Generation Purple flowers Appearance: Genetic makeup: PP Gametes: White flowers pp p P F 1 Generation Appearance: Genetic makeup: Gametes: Purple flowers Pp 1/ 2 P Sperm F 2 Generation P p PP Pp Pp pp P Eggs p 3 1 p

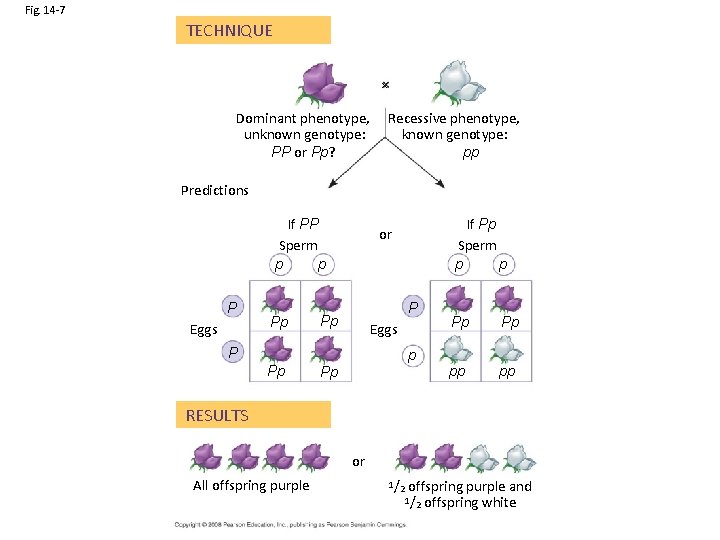
Fig. 14 -7 TECHNIQUE Dominant phenotype, unknown genotype: PP or Pp? Recessive phenotype, known genotype: pp Predictions If PP Sperm p p P Eggs Pp If Pp Sperm p p or P Pp Eggs P Pp p Pp Pp Pp pp pp RESULTS or All offspring purple 1/2 offspring purple and offspring white 1/2

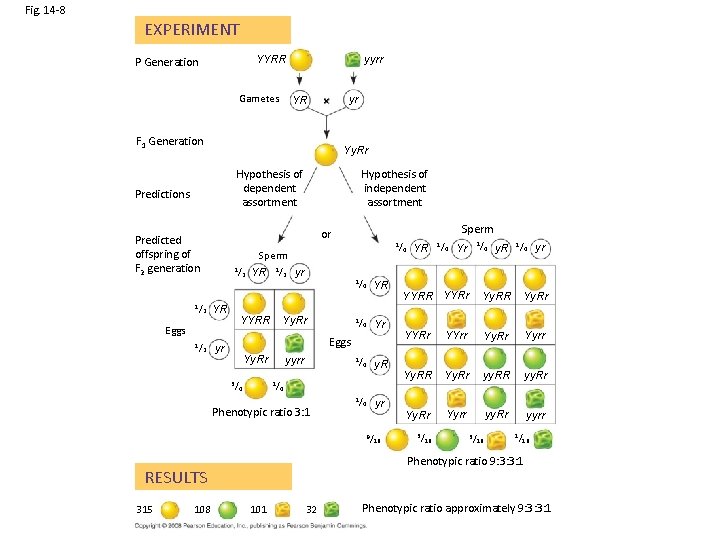
Fig. 14 -8 EXPERIMENT YYRR P Generation Gametes yyrr YR F 1 Generation yr Yy. Rr Hypothesis of dependent assortment Predictions 1/ Sperm 1/ 2 YR 2 2 YR 1/ 2 yr 1/ Yy. Rr YYRR Eggs 1/ Sperm or Predicted offspring of F 2 generation 1/ Hypothesis of independent assortment 1/ 4 4 YR Yr Eggs yr Yy. Rr 3/ yyrr 1/ 4 1/ y. R 4 1/ yr 4 9/ 4 1/ Yr 4 y. R 1/ 4 yr YYRR YYRr Yy. RR Yy. Rr YYrr Yy. Rr Yyrr Yy. RR Yy. Rr yy. RR yy. Rr Yyrr yy. Rr yyrr 16 3/ 16 1/ 16 Phenotypic ratio 9: 3: 3: 1 RESULTS 108 YR 4 Phenotypic ratio 3: 1 315 4 1/ 101 32 Phenotypic ratio approximately 9: 3: 3: 1

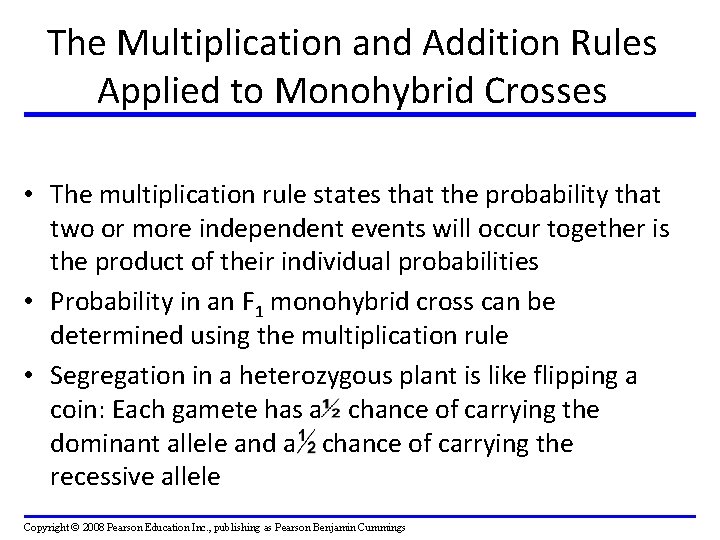
The Multiplication and Addition Rules Applied to Monohybrid Crosses • The multiplication rule states that the probability that two or more independent events will occur together is the product of their individual probabilities • Probability in an F 1 monohybrid cross can be determined using the multiplication rule • Segregation in a heterozygous plant is like flipping a coin: Each gamete has a chance of carrying the dominant allele and a chance of carrying the recessive allele Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

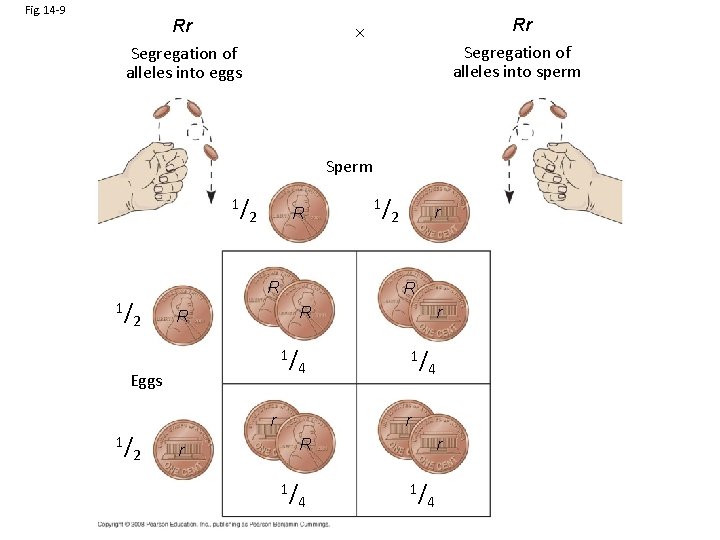
Fig. 14 -9 Rr Segregation of alleles into eggs Rr Segregation of alleles into sperm Sperm 1/ R 2 R 1/ r 1/ 4 r 2 R R Eggs 1/ 1/ 4 r r R r 1/ 4

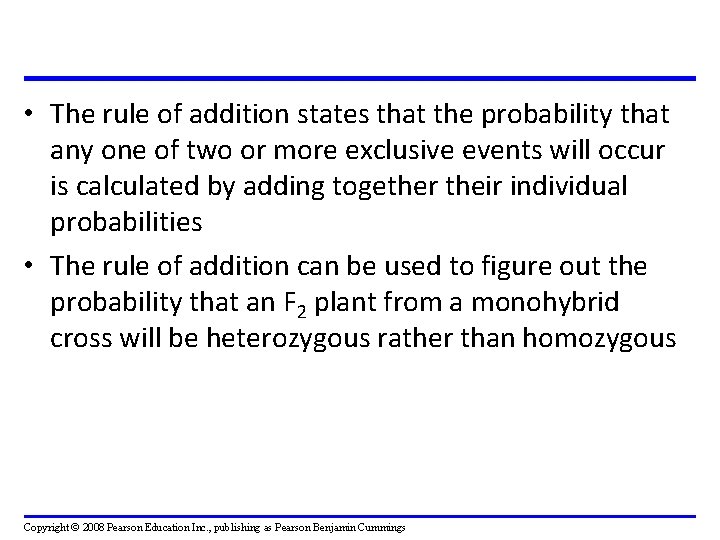
• The rule of addition states that the probability that any one of two or more exclusive events will occur is calculated by adding together their individual probabilities • The rule of addition can be used to figure out the probability that an F 2 plant from a monohybrid cross will be heterozygous rather than homozygous Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

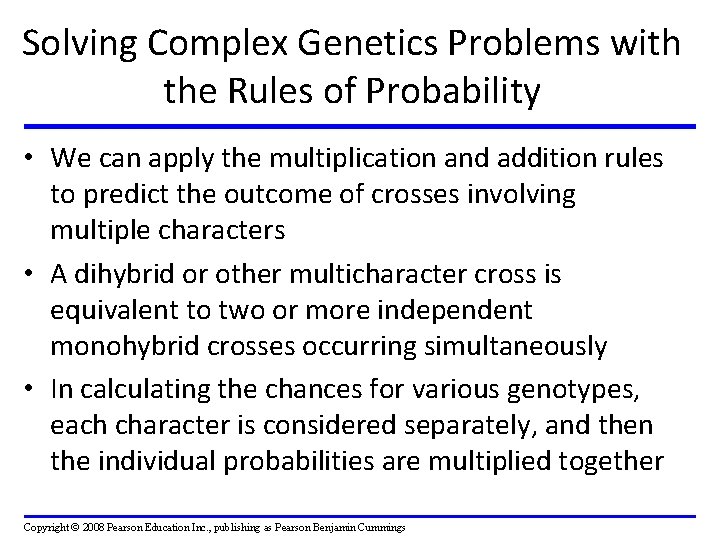
Solving Complex Genetics Problems with the Rules of Probability • We can apply the multiplication and addition rules to predict the outcome of crosses involving multiple characters • A dihybrid or other multicharacter cross is equivalent to two or more independent monohybrid crosses occurring simultaneously • In calculating the chances for various genotypes, each character is considered separately, and then the individual probabilities are multiplied together Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

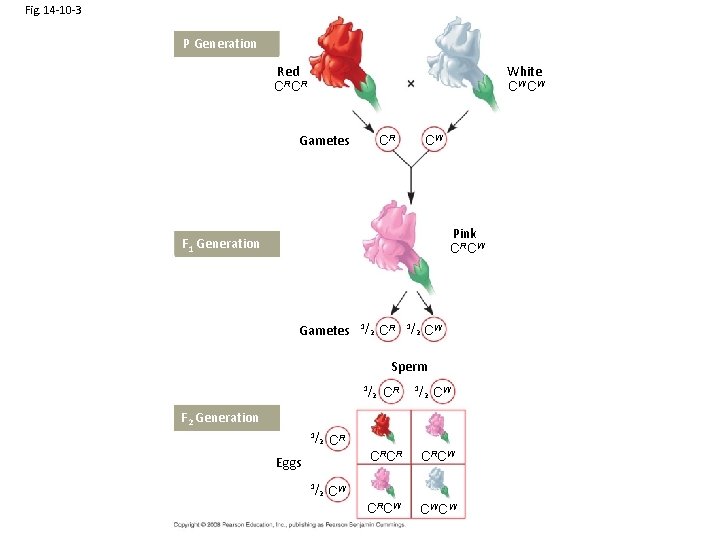
Fig. 14 -10 -3 P Generation Red CRCR White CW CW Gametes CR CW Pink CRCW F 1 Generation Gametes 1/ 2 CR 1/ 2 CW Sperm 1/ 2 CR 1/ 2 CW F 2 Generation 1/ 2 CR Eggs 1/ 2 CRCR CRCW CW

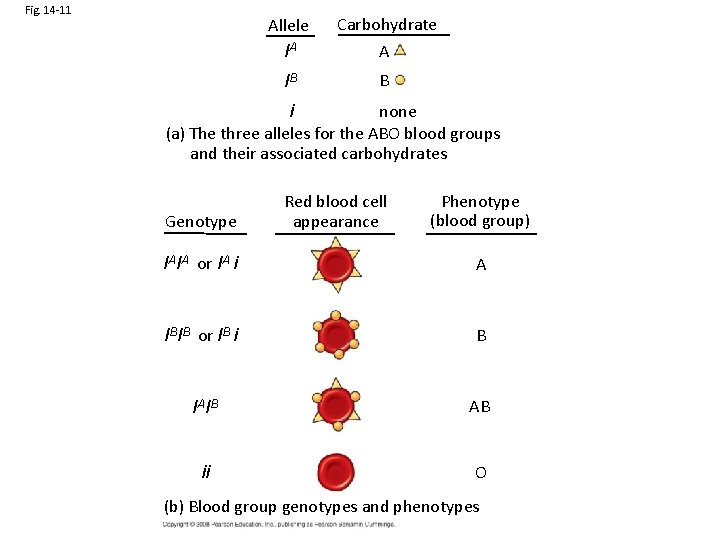
Fig. 14 -11 Allele IA Carbohydrate A IB B none i (a) The three alleles for the ABO blood groups and their associated carbohydrates Genotype Red blood cell appearance Phenotype (blood group) IAIA or IA i A IBIB or IB i B IAIB AB ii O (b) Blood group genotypes and phenotypes

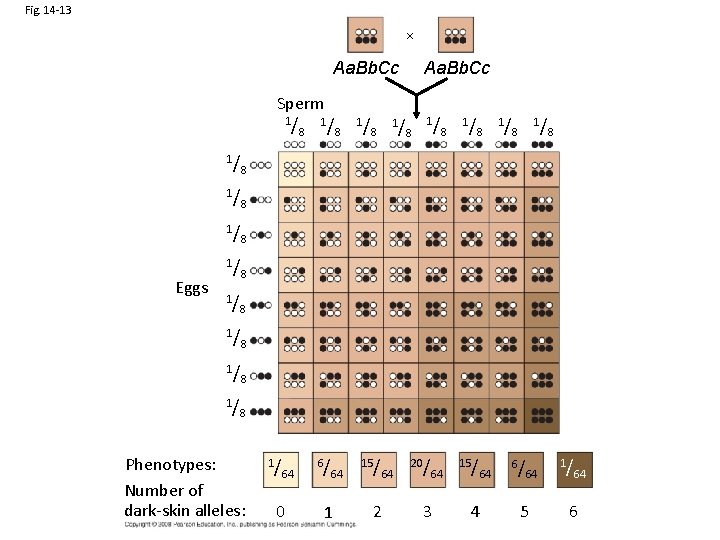
Fig. 14 -13 Aa. Bb. Cc Sperm 1/ Eggs 1/ 8 1/ 1/ 8 8 1/ 64 15/ 8 1/ 1/ 8 8 8 1/ 8 Aa. Bb. Cc 8 Phenotypes: Number of dark-skin alleles: 1/ 64 0 6/ 64 1 15/ 64 2 20/ 3 64 4 6/ 64 5 1/ 64 6

Fig. 14 -14

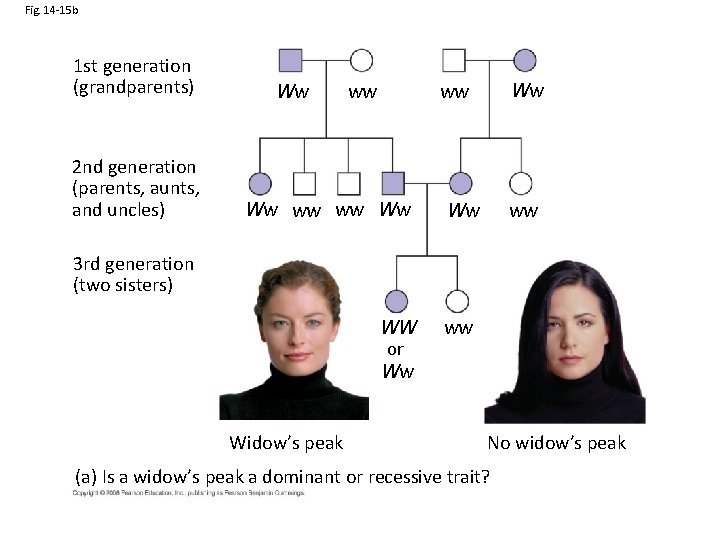
Fig. 14 -15 b 1 st generation (grandparents) 2 nd generation (parents, aunts, and uncles) Ww ww ww Ww Ww ww 3 rd generation (two sisters) WW or Ww Widow’s peak ww No widow’s peak (a) Is a widow’s peak a dominant or recessive trait?

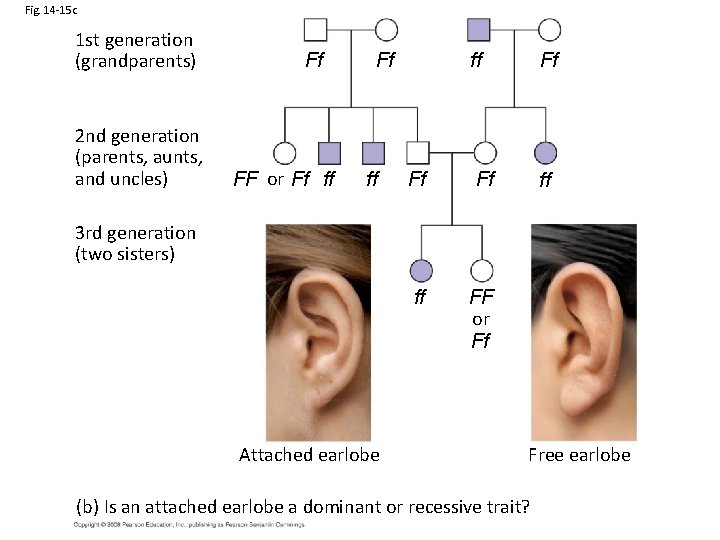
Fig. 14 -15 c 1 st generation (grandparents) 2 nd generation (parents, aunts, and uncles) Ff FF or Ff ff ff Ff Ff Ff ff ff FF or Ff 3 rd generation (two sisters) Attached earlobe Free earlobe (b) Is an attached earlobe a dominant or recessive trait?

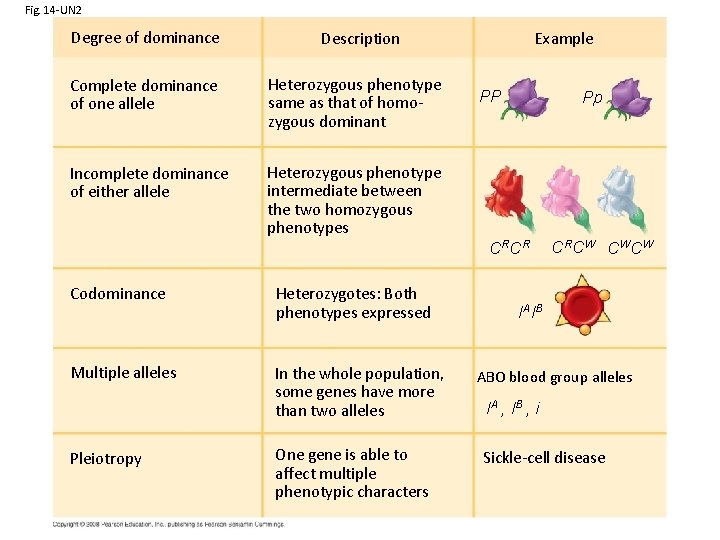
Fig. 14 -UN 2 Degree of dominance Example Description Complete dominance of one allele Heterozygous phenotype same as that of homozygous dominant Incomplete dominance of either allele Heterozygous phenotype intermediate between the two homozygous phenotypes PP Pp CR CR Codominance Heterozygotes: Both phenotypes expressed Multiple alleles In the whole population, some genes have more than two alleles Pleiotropy One gene is able to affect multiple phenotypic characters CR CW CW CW IA IB ABO blood group alleles IA , I B , i Sickle-cell disease

Chromosomal Theory of Inheritance

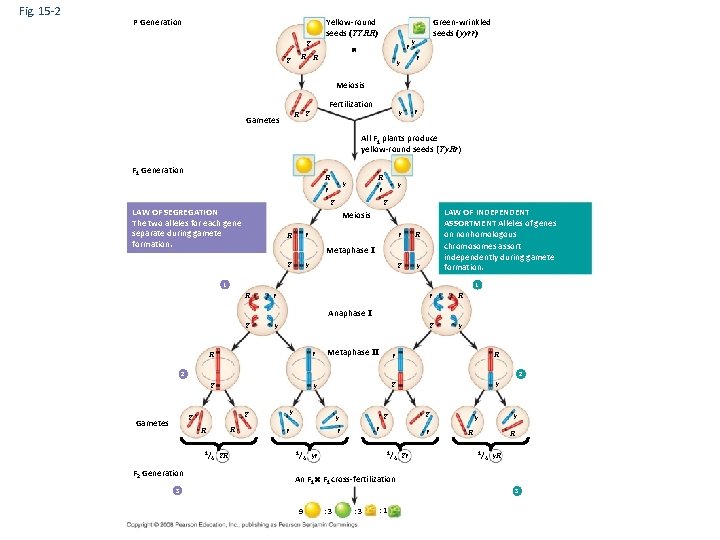
Fig. 15 -2 P Generation Yellow-round seeds (YYRR) Y Y R r R y Green-wrinkled seeds ( yyrr) y r Meiosis Fertilization y R Y Gametes r All F 1 plants produce yellow-round seeds (Yy. Rr) F 1 Generation R R y r Y Y LAW OF SEGREGATION The two alleles for each gene separate during gamete formation. y r LAW OF INDEPENDENT ASSORTMENT Alleles of genes on nonhomologous chromosomes assort independently during gamete formation. Meiosis R r Y y r R Y y Metaphase I 1 1 R r Y y r R Y y Anaphase I R r Y y Metaphase II r R Y y 2 2 Y Y Gametes R R 1/ F 2 Generation 4 YR y r r r 1/ Y Y y r 1/ 4 yr 4 Yr y y R R 1/ 4 y. R An F 1 cross-fertilization 3 3 9 : 3 : 1

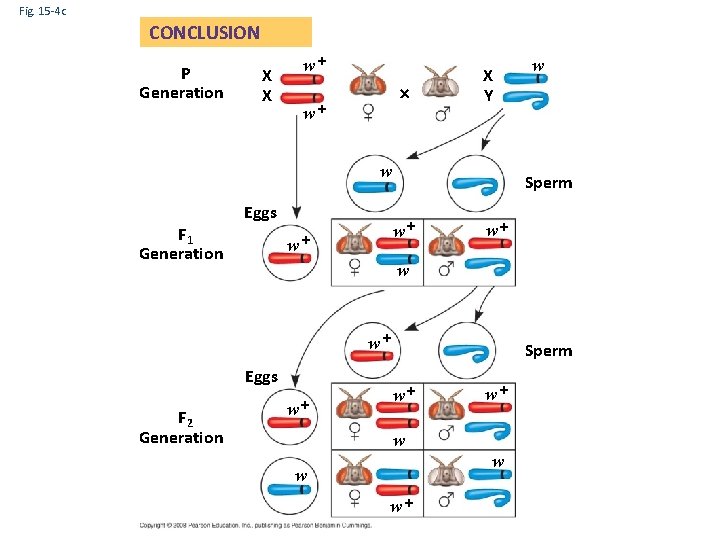
Fig. 15 -4 c CONCLUSION P Generation X X w+ w+ X Y w F 1 Generation Eggs Sperm w+ w+ w+ w w+ Eggs F 2 Generation w w+ Sperm w+ w+ w w+

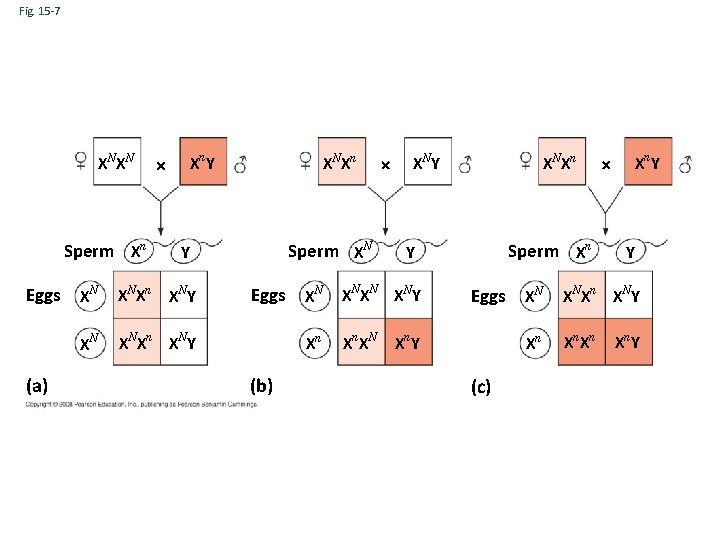
Fig. 15 -7 XN XN Sperm Xn Xn Y Sperm XN Y Eggs XN XN Xn XN Y (a) XN Xn Eggs (b) XN Y XN Xn Sperm Xn Y XN XN Y Xn Xn XN Eggs XN Xn Y Xn (c) Xn Y Y XN Xn XN Y Xn Xn Xn Y

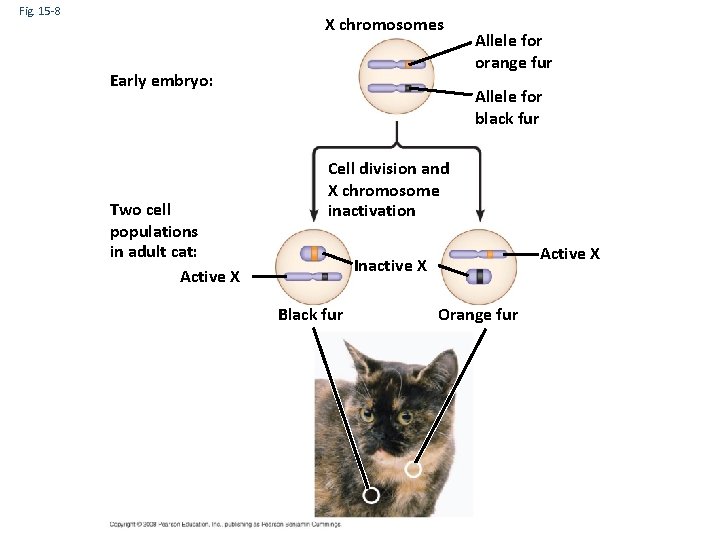
Fig. 15 -8 X chromosomes Early embryo: Two cell populations in adult cat: Active X Allele for orange fur Allele for black fur Cell division and X chromosome inactivation Active X Inactive X Black fur Orange fur

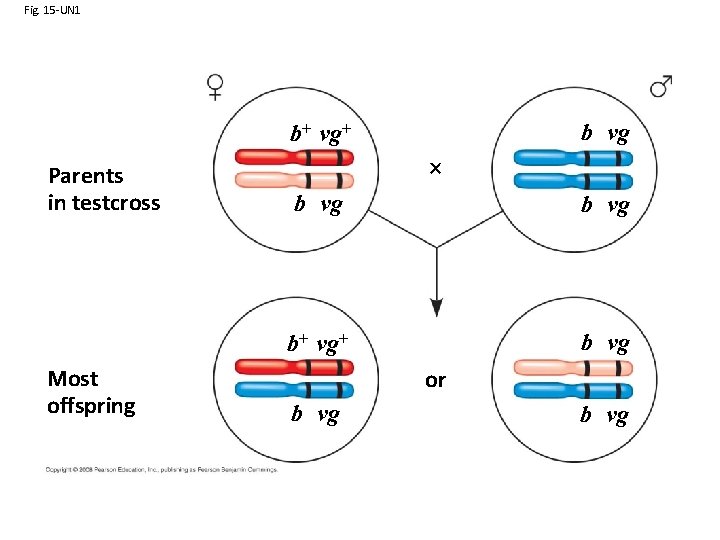
Fig. 15 -UN 1 b vg b+ vg+ Parents in testcross Most offspring b vg b+ vg+ b vg or b vg

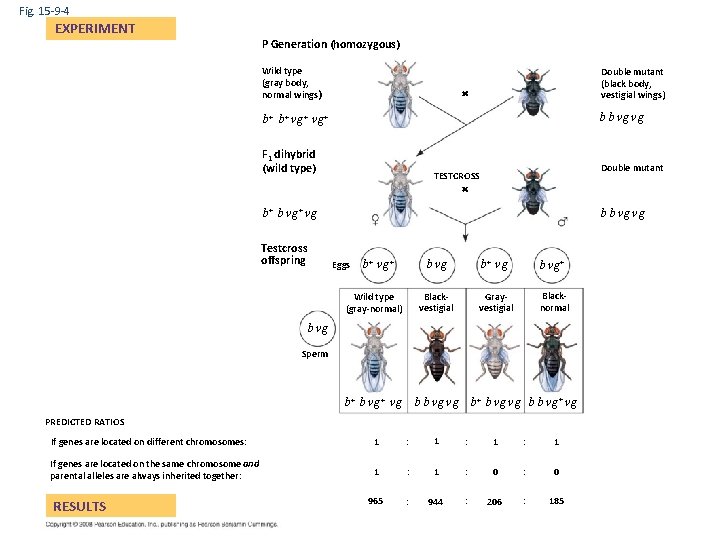
Fig. 15 -9 -4 EXPERIMENT P Generation (homozygous) Wild type (gray body, normal wings) Double mutant (black body, vestigial wings) b b vg vg b+ b+ vg+ F 1 dihybrid (wild type) Double mutant TESTCROSS b+ b vg+ vg Testcross offspring b b vg vg Eggs b+ vg+ Wild type (gray-normal) b vg b+ vg b vg+ Blackvestigial Grayvestigial Blacknormal b vg Sperm b b vg vg b+ b vg vg b b vg+ vg b+ b vg+ vg PREDICTED RATIOS If genes are located on different chromosomes: 1 If genes are located on the same chromosome and parental alleles are always inherited together: 1 : 0 965 : 944 : 206 : 185 RESULTS

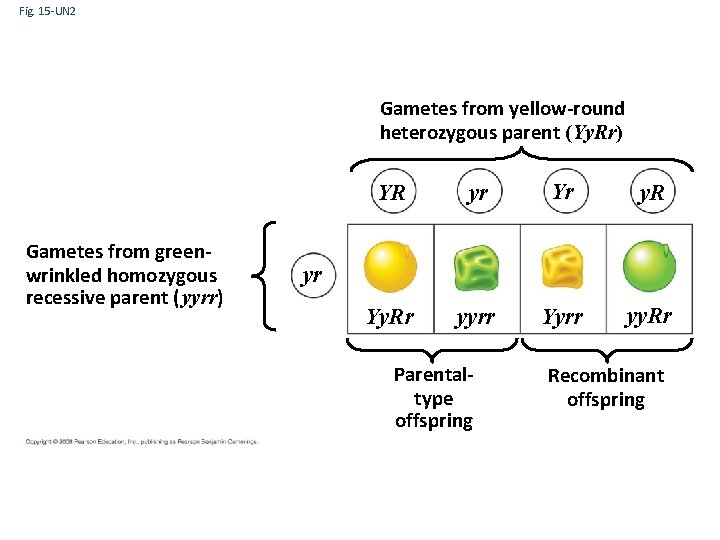
Fig. 15 -UN 2 Gametes from yellow-round heterozygous parent (Yy. Rr) Gametes from greenwrinkled homozygous recessive parent ( yyrr) YR yr Yr y. R Yy. Rr yyrr Yyrr yy. Rr yr Parentaltype offspring Recombinant offspring

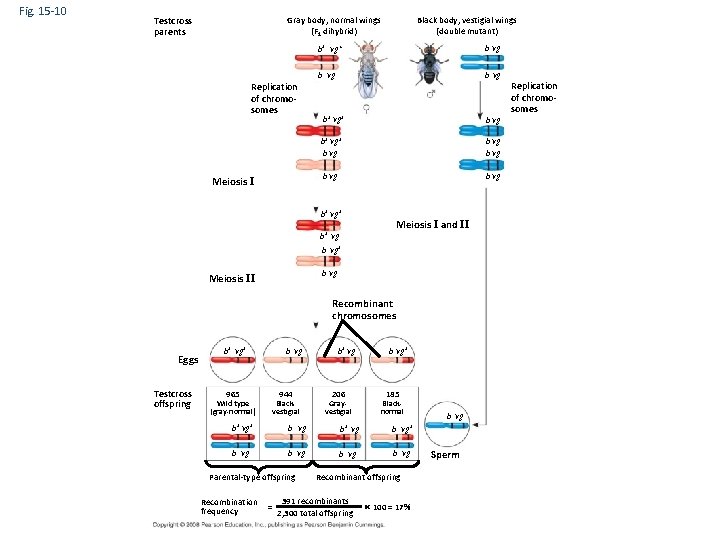
Fig. 15 -10 Testcross parents Gray body, normal wings (F 1 dihybrid) Replication of chromosomes Meiosis I Black body, vestigial wings (double mutant) b+ vg+ b vg Replication of chromosomes b+ vg+ b vg b vg b+ vg+ b+ vg Meiosis I and II b vg+ b vg Meiosis II Recombinant chromosomes Eggs Testcross offspring b vg b+ vg+ 965 944 Wild type (gray-normal) Blackvestigial b+ vg 206 Grayvestigial b vg+ 185 Blacknormal b+ vg+ b vg b+ vg b vg+ b vg Parental-type offspring Recombination frequency = Recombinant offspring 391 recombinants 2, 300 total offspring 100 = 17% b vg Sperm

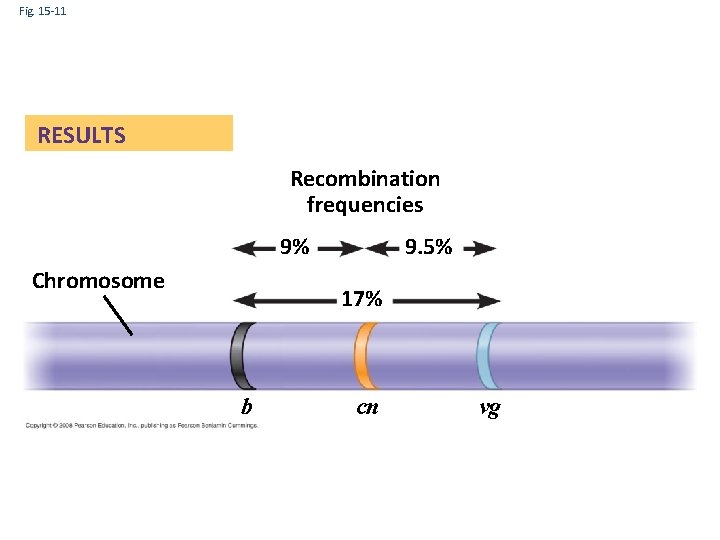
Fig. 15 -11 RESULTS Recombination frequencies 9% Chromosome 9. 5% 17% b cn vg

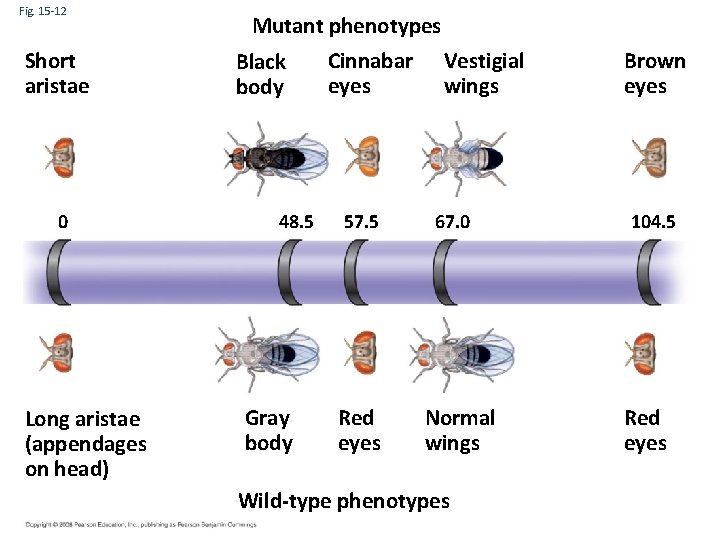
Fig. 15 -12 Short aristae 0 Long aristae (appendages on head) Mutant phenotypes Black body 48. 5 Gray body Cinnabar eyes 57. 5 Red eyes Vestigial wings 67. 0 Normal wings Wild-type phenotypes Brown eyes 104. 5 Red eyes

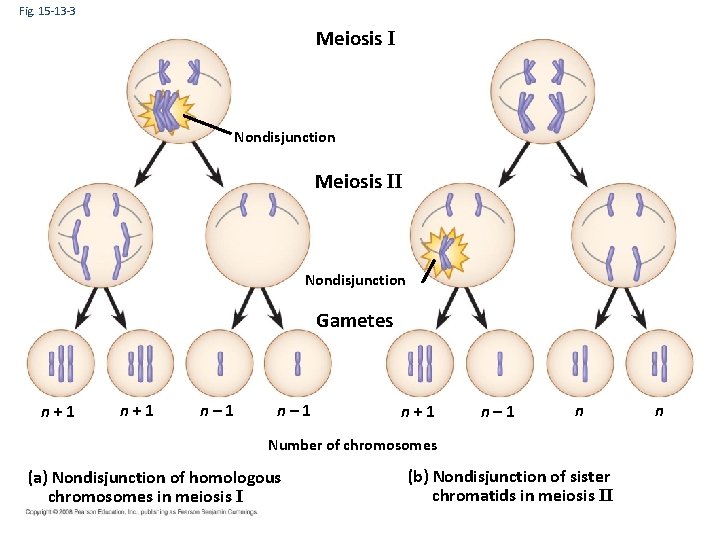
Fig. 15 -13 -3 Meiosis I Nondisjunction Meiosis II Nondisjunction Gametes n+1 n– 1 n Number of chromosomes (a) Nondisjunction of homologous chromosomes in meiosis I (b) Nondisjunction of sister chromatids in meiosis II n

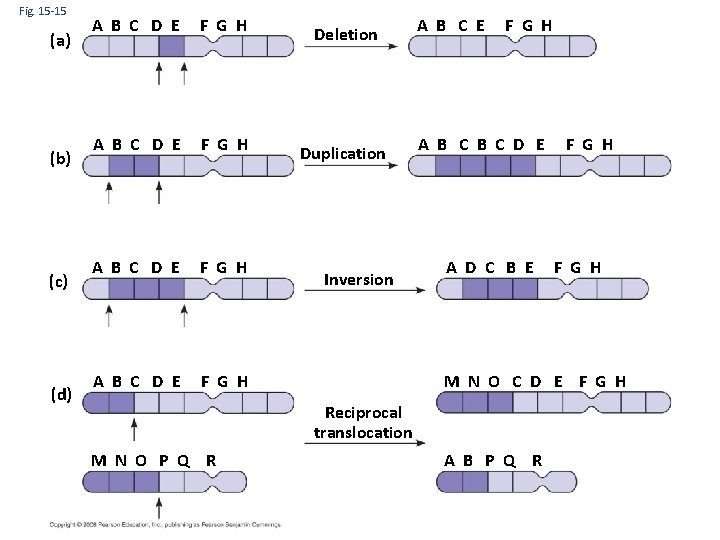
Fig. 15 -15 (a) (b) (c) (d) A B C D E F G H Deletion Duplication Inversion A B C E F G H A B C D E A D C B E F G H M N O C D E F G H Reciprocal translocation M N O P Q R F G H A B P Q R

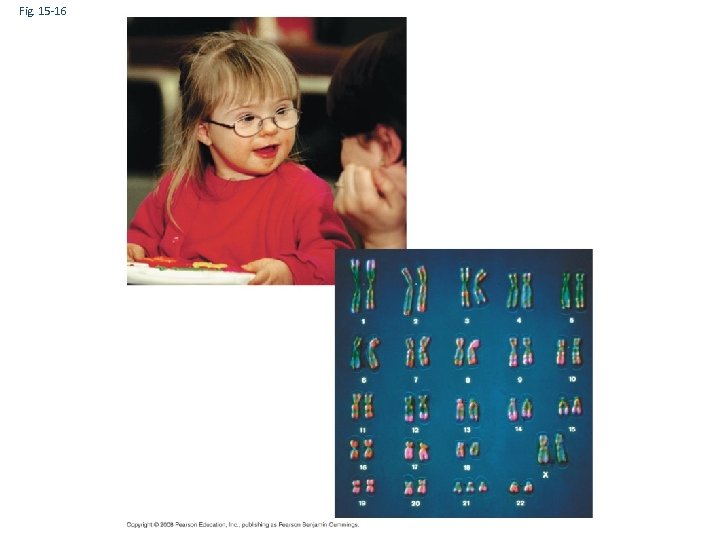
Fig. 15 -16

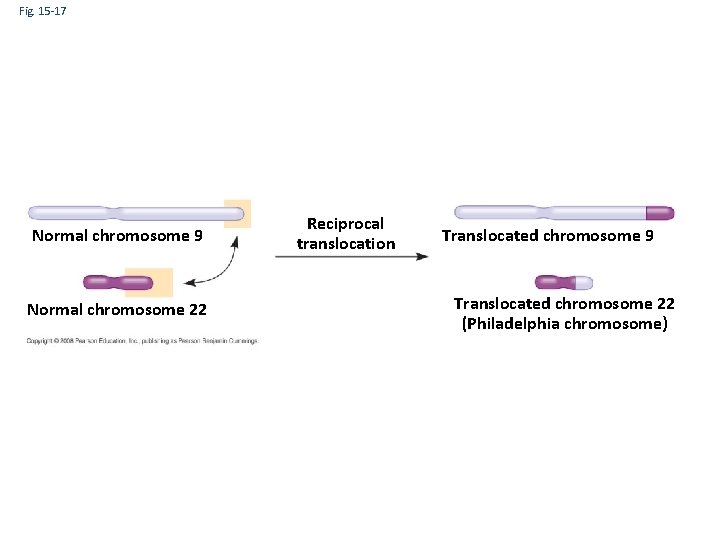
Fig. 15 -17 Normal chromosome 9 Normal chromosome 22 Reciprocal translocation Translocated chromosome 9 Translocated chromosome 22 (Philadelphia chromosome)