Microfluidics for Gene Fabrication Peter Carr David Kong
Microfluidics for Gene Fabrication Peter Carr & David Kong MIT Media Laboratory
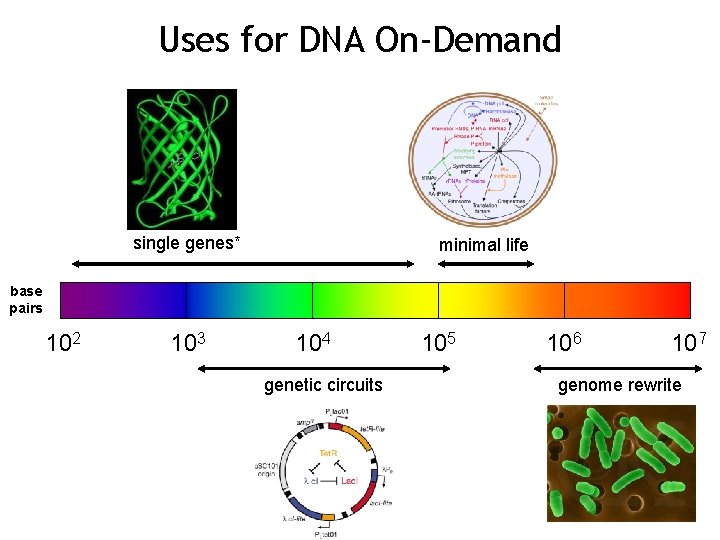
Uses for DNA On-Demand single genes* minimal life base pairs 102 103 104 genetic circuits 105 106 107 genome rewrite
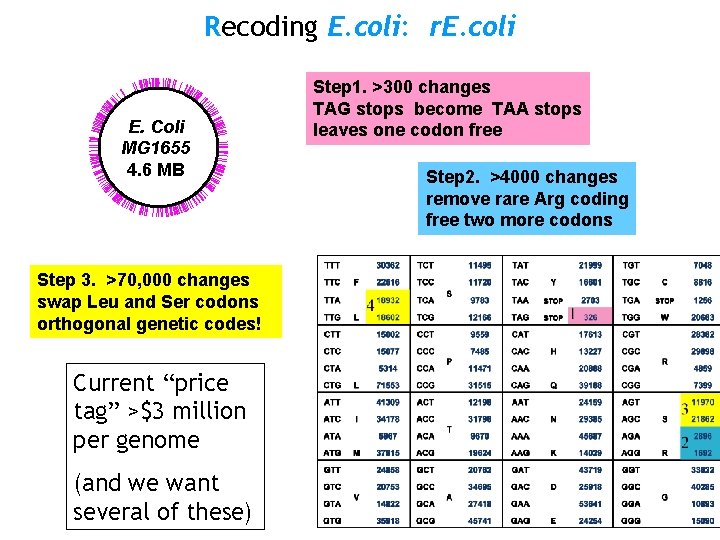
Recoding E. coli: r. E. coli E. Coli MG 1655 4. 6 MB Step 3. >70, 000 changes swap Leu and Ser codons orthogonal genetic codes! Current “price tag” >$3 million per genome (and we want several of these) Step 1. >300 changes TAG stops become TAA stops leaves one codon free Step 2. >4000 changes remove rare Arg coding free two more codons

Trends in de novo DNA synthesis Carlson, R. (2003) The Pace and Proliferation of Biological Technologies

SIRV-1: hjc gene parse 25 nt Primers: t 1 = b 16 = 25 nt Construction oligos: b 2 t 15 = 48 nt total gene: = 390 bp 25 nt t 1 t 3 t 5 CAGGTAATTCCATATGAACATCCGTCAGTCTGGTAAATACTACGAGTACAAAACTCTGGAGATCCTGGAAAAGAATGGTTTCAAAGCGCTGCGTATCCCGGTTTCTGGTACCGGCAAACAGGCGCTGCC GTCCATTAAGGTATACTTGTAGGCAGTCAGACCATTTATGATGCTCATGTTTTGAGACCTCTAGGACCTTTTCTTACCAAAGTTTCGCGACGCATAGGGCCAAAGACCATGGCCGTTTGTCCGCGACGG CCATTAAGGTATACTTGTAGGCAGTCAGACCATTTATGATGCTCATGT CTAGGACCTTTTCTTACCAAAGTTTCGCGACGCATAGGG CGTTTGTCCGCGACGG b 2 b 4 t 7 t 9 GGACCTGATCGCGACCAAAAACAACACCATCTACCCTATTGAAGTTAAATCTACCTCTAAAGACGTTGTTACCGTTCGTAATTTCCAGATCGAAAAACTGTTCAAATTCTGCGCGAAATCTTCAACTTCTGTGA CGACCAAAAACAACACCATCTACCCTATTGAAGTTAAATCTACCTCTA GACGTTGTTACCGTTCGTAATTTCCAGATCGAAAAACTGTTCAAATT GCGAAATCTTCAACTTCTGTGA CCTGGACTAGCGCTGGTTTTTGTTGTGGTAGATGGGATAACTTCAATTTAGATGGAGATTTCTGCAACAATGGCAAGCATTAAAGGTCTAGCTTTTTGACAAGTTTAAGACGCTTTAGAAGTTGAAGACACTCT CCTGGACTAGCGCTGGTTTTTGTTGTGGTAGA GGGATAACTTCAATTTAGATGGAGATTTCTGCAACAATGGCAAGCATT GGTCTAGCTTTTTGACAAGTTTAAGACGCTTTAGAAGTTGAAGACACT b 6 b 8 t 11 b 10 t 13 t 15 ATGCCACCCGCTGGTAACCGTTTACTACAAGAAATACAAAATCGTTTATGAACTGTCTCAGGACGTTCGCACCAAAGAAAAAATCAAGTACGGCATCAACTCCTAACTCGAGCGGAC ATGCCACCCGCTGGTAACCGTTTACT CAAGAAATACAAAATCGTTTATGAACTGTCTCAGGACGTTCG CCAAAGAAAAAATCAAGTACGGCATCAACTCCTAACTCGAGC TACGGTGGGCGACCATTGGCAAATGATGTTCTTTATGTTTTAGCAAATACTTGACAGAGTCCTGCAAGCGTGGTTTCTTTTTTAGTTCATGCCGTAGTTGAGGATTGAGCTCGCCTG GGCGACCATTGGCAAATGATGTTCTTTATGTTTTAGCAAATACTTGACAGAGTCCTGCAAGCGTGGTTTCTTTTTTAGTTCAAGTT CGTAGTTGAGGATTGAGCTCGCCTG b 12 b 14 b 16

SIRV-1: hjc one-step PCA t 1 t 3 b 2 t 5 b 4 t 7 b 6 t 9 b 8 50 bp t 11 b 10 t 13 t 15 b 12 b 14 73 bp 108 bp 390 bp b 16
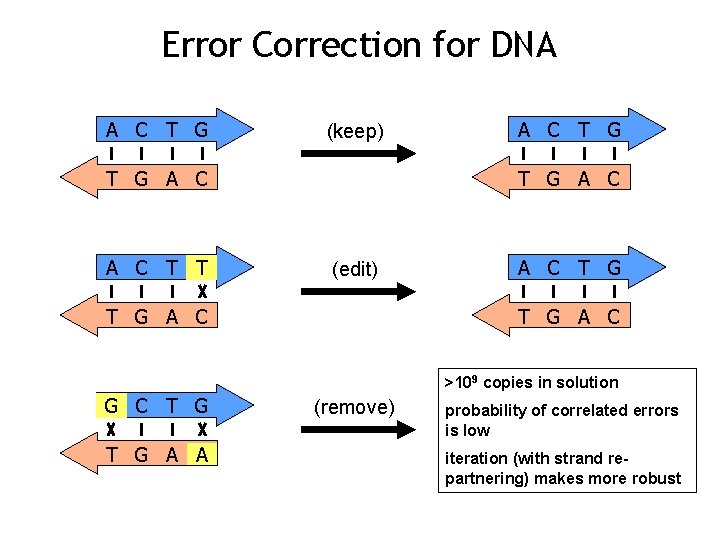
Error Correction for DNA A C T G (keep) T G A C T T A C T G A C (edit) T G A C >109 copies in solution G C T G A A (remove) probability of correlated errors is low iteration (with strand repartnering) makes more robust

DNA Error Correction Protocols Hybridization-selection Mismatch Binding/Removal Mismatch Cleavage bind Mut. S remove Mut. S + mismatch (error-free DNA) Tian et al. Nature 432 (2004) Carr et al. NAR 32 (2004) Smith & Modrich PNAS 94 (1997)

High Density Oligonucleotide Microarrays a massive feedstock of DNA building blocks inkjet-printed microarrays (e. g. Agilent) maskless array synthesizer (e. g. Nimblegen) >105 oligos per >5 megabases >1000 x reduction in = of DNA information = oligonucleotide costs microarray Tian & Church (2004): ~600 oligos 21 genes 15 kb construct

Making it Fast & Cheap: Challenges • Harnessing the full potential of oligo microarrays – minute amounts of material: amplification? – assembly: erratic behaviors of increasingly complex mixes • Minimizing expensive reagents (polymerases, error correction) • High throughput parallel sample handling • Process integration

Microfluidic Gene Synthesis: Integration Road Map oligo microarray synthesis user designs DNA error correction clone sequence/QC assemble DNA constructs larger scale assembly express/assay DATA (MOLECULES) DATA

Parallel microfluidic gene synthesis Fabrication PDMS 1 PDMS 2 PDMS 3 CYTOP, Paralyne, Four parallel 500 -n. L reactors

Parallel microfluidic gene synthesis Gene Total size (nt) Number of oligos Construction Oligo size (nt) Amplifying Primer sizes (nt) alba 327 16 38 35, 32 hjc 390 16 48 25, 25 ble 461 16 47 31, 45 Ds. Red 733 26 50 25, 20 OR 128 -1 942 32 50 31, 37 GFP 993 42 42 29, 29 Kong et al. Nucleic Acids Research, 2007

Parallel microfluidic gene synthesis Sequencing results Error Type Deletion Single-base 1 in 600 Microfluidic Device PCR tube 19 16 1 in 1400 1 in 10, 00 Deletion Multiple-base 5 5 Transition G/C to A/T 3 6 Transition A/T to G/C 0 2 Transversion G/C to C/G 0 0 Transversion G/C to T/A 1 1 Transversion A/T to C/G 1 0 Transversion A/T to T/A 0 0 Total Errors: 29 30 Bases Sequenced: 16, 250 13, 389 Error Rate (per base): 0. 0018 0. 0022 Carr et al. Nucleic Acids Research, 2004 -verified identity of each gene by sequencing -12. 5% of clones were error-free in agreement with theoretical predictions Kong et al. Nucleic Acids Research, 2007

Microfluidic Gene Synthesis: Integration Road Map oligo microarray synthesis user designs DNA error correction clone sequence/QC assemble DNA constructs larger scale assembly express/assay DATA (MOLECULES) DATA

Integrated Microarray-Microfluidics -Perform synthesis without pre-assembly amplification -Enables increased utilization of high-density DNA microarrays by: -reducing pool complexity -limiting undesired oligo interactions -maintaining reagent concentrations at desired levels Agilent Nimblegen (MAS) Oligo spot feature size 135 m 16 m Area of spot 1. 4 x 104 m 2 2. 56 x 102 m 2 Oligo density 0. 1 picomole/mm 2 Maximum expected yield per spot 1. 4 femtomole 0. 0256 femtomole Dimensions of spot spacing 212 m by 188 m 25 μm by 25 μm Minimal footprint of 16 oligo spots 6. 4 x 105 m 2 1 x 104 m 2 Minimal chamber volume (10 m height) 6. 4 nanoliters 100 picoliters Estimated concentration of each oligo 220 n. M 256 n. M

Microfluidic Gene Synthesis: Integration Road Map oligo microarray synthesis user designs DNA error correction clone sequence/QC assemble DNA constructs larger scale assembly express/assay DATA (MOLECULES) DATA

Hierarchical gene synthesis

Hierarchical gene synthesis E • 300 n. L reactors • On-chip mixing of synthesized fragments A, B, with a “rejuvenating mixture” of d. NTPs, polymerase, and amplifying primers • Fragment A, B, and E sample collection (real time)

Microfluidic Gene Synthesis: Integration Road Map oligo microarray synthesis user designs DNA error correction clone sequence/QC assemble DNA constructs larger scale assembly express/assay DATA (MOLECULES) DATA

Integrated gene and protein synthesis oligos gene protein Fluorescence from GFP expressed in a PDMS microfluidic device 45 n. L gene synthesis reactors, 12 n. L protein synthesis reactors Fluorescence from expressed synthetic EGFP

Thank You Lu Chen Kelly Chang Byron Hsu Dr. Shuguang Zhang Prof. George Church Prof. Franco Cerrina Prof. Joseph Jacobson Funding: MIT Media Lab Center for Bits and Atoms (NSF)
- Slides: 22