Micrococcus luteus on blood agar A microbiologists view

Micrococcus luteus on blood agar
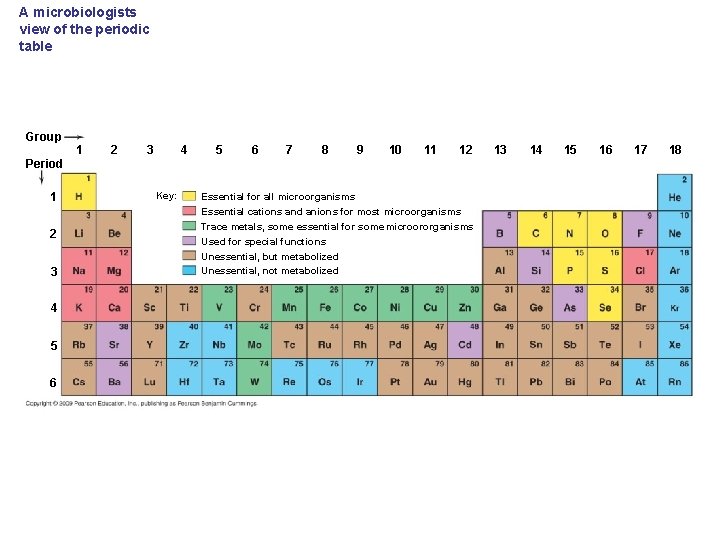
A microbiologists view of the periodic table Group 1 2 4 3 5 6 7 8 9 10 11 12 Period 1 2 3 4 5 6 Key: Essential for all microorganisms Essential cations and anions for most microorganisms Trace metals, some essential for some microororganisms Used for special functions Unessential, but metabolized Unessential, not metabolized 13 14 15 16 17 18
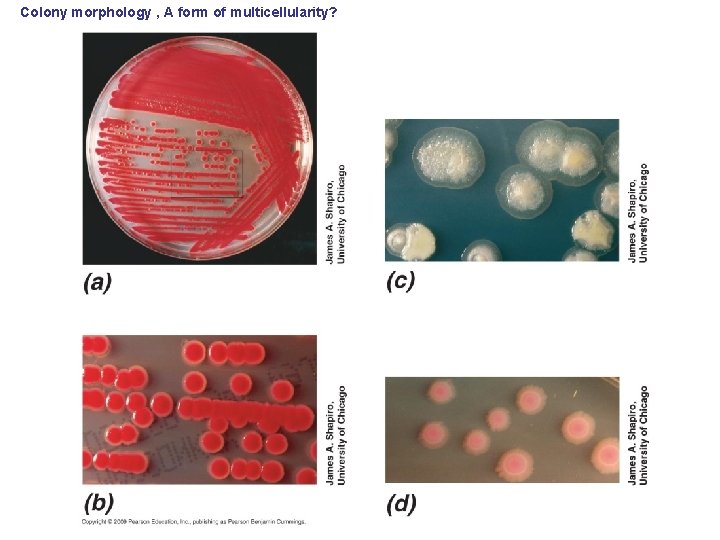
Colony morphology , A form of multicellularity?
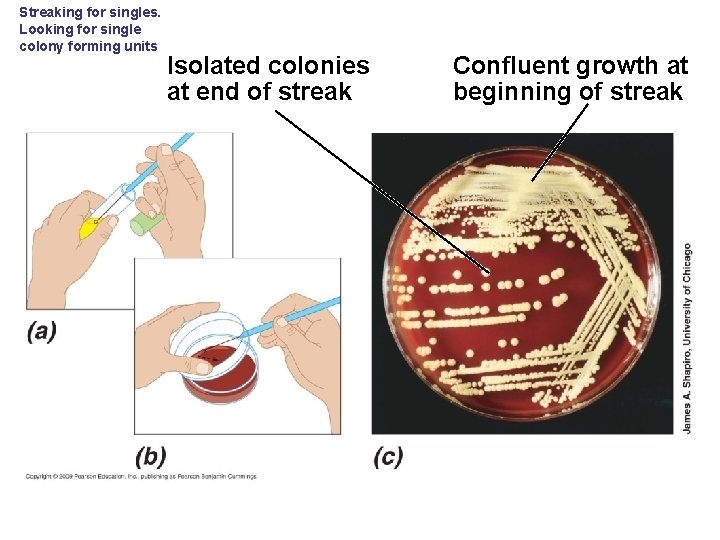
Streaking for singles. Looking for single colony forming units Isolated colonies at end of streak Confluent growth at beginning of streak
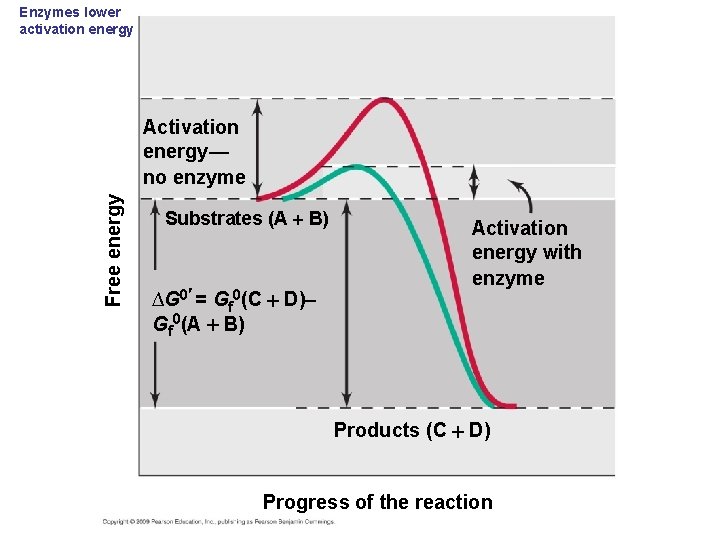
Enzymes lower activation energy Free energy Activation energy— no enzyme Substrates (A B) ∆G 0 = Gf 0(C D) Gf 0(A B) Activation energy with enzyme Products (C D) Progress of the reaction
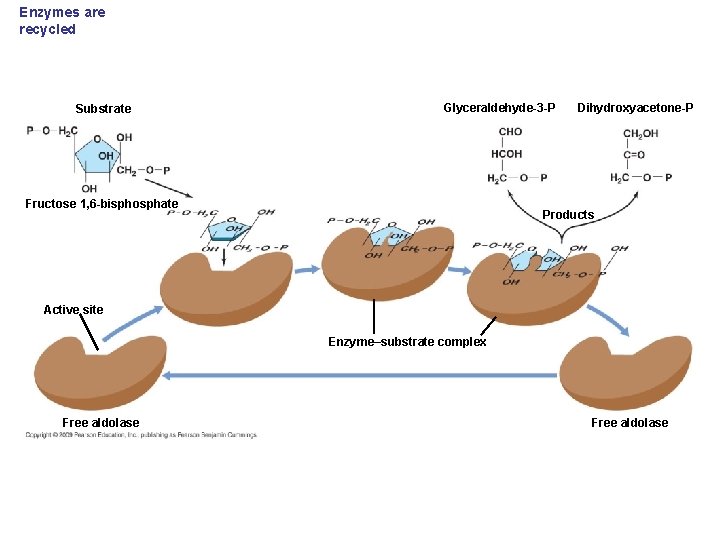
Enzymes are recycled Substrate Glyceraldehyde-3 -P Fructose 1, 6 -bisphosphate Dihydroxyacetone-P Products Active site Enzyme–substrate complex Free aldolase

Enzymes are specific for their substrates 3 dimensional structure determined by folding is dependent on side chain interactions determined by charge and hydrophobicity.
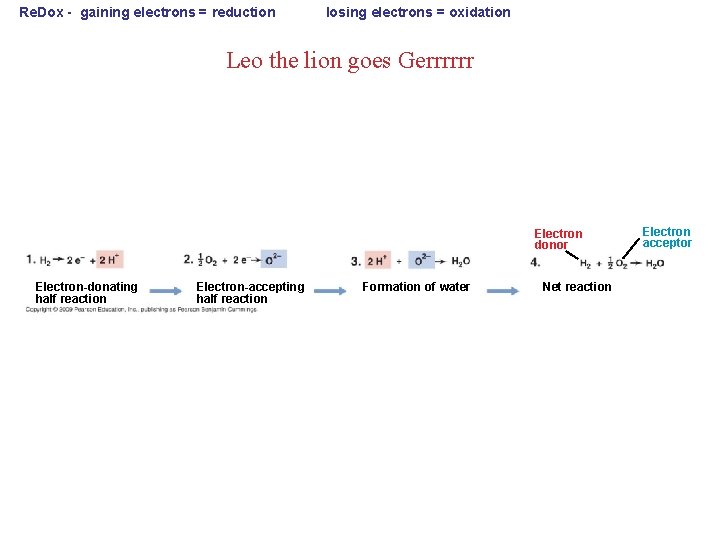
Re. Dox - gaining electrons = reduction losing electrons = oxidation Leo the lion goes Gerrrrrr Electron donor Electron-donating half reaction Electron-accepting half reaction Formation of water Net reaction Electron acceptor
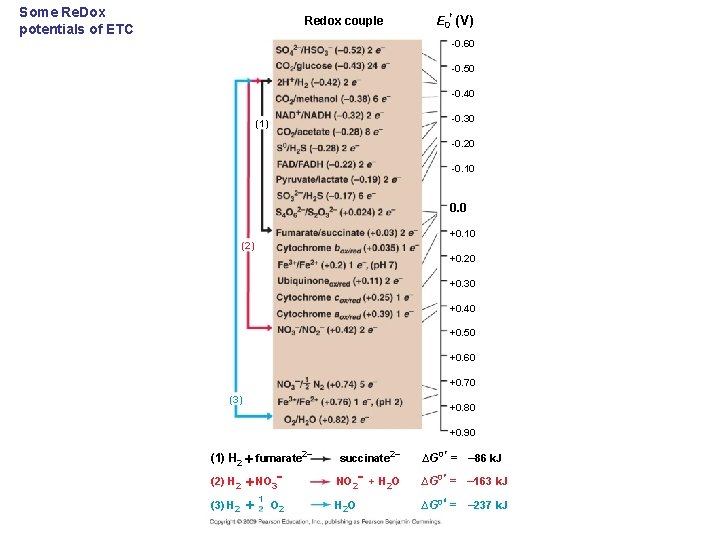
Some Re. Dox potentials of ETC E 0 (V) Redox couple -0. 60 -0. 50 -0. 40 -0. 30 (1) -0. 20 -0. 10 0. 0 +0. 10 (2) +0. 20 +0. 30 +0. 40 +0. 50 +0. 60 +0. 70 (3) +0. 80 +0. 90 (1) H 2 fumarate 2 succinate (2) H 2 NO 3 NO 2 (3) H 2 O O 2 2 + H 2 O ∆G 0 = – 86 k. J ∆G 0 = – 163 k. J ∆G 0 = – 237 k. J
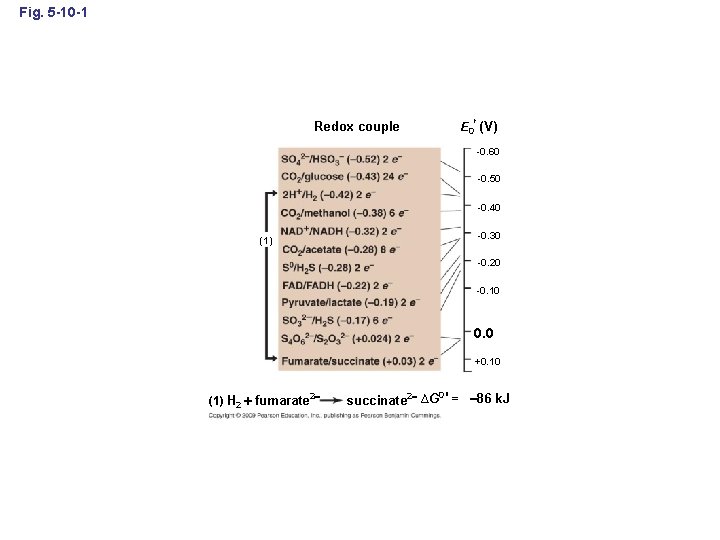
Fig. 5 -10 -1 Redox couple E 0 (V) -0. 60 -0. 50 -0. 40 (1) -0. 30 -0. 20 -0. 10 0. 0 +0. 10 (1) H 2 fumarate 2 0 succinate 2 ∆G = – 86 k. J
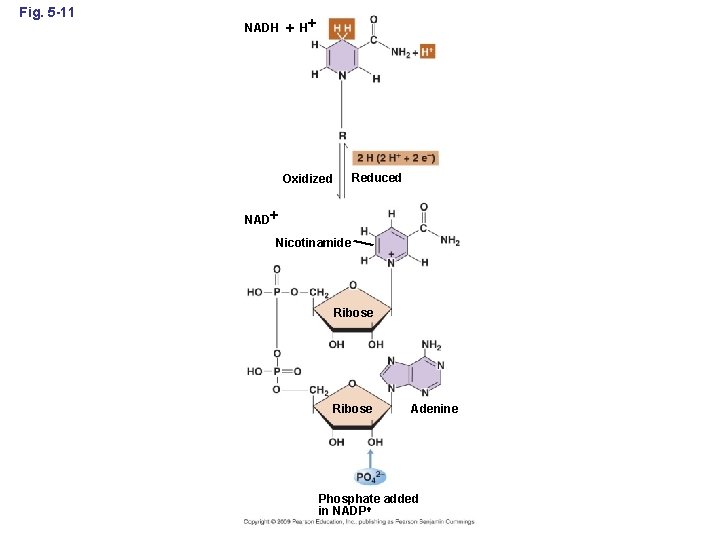
Fig. 5 -11 NADH H Reduced Oxidized NAD Nicotinamide Ribose Adenine Phosphate added in NADP

Fig. 5 -12 Reaction 1. Enzyme I reacts with electron donor and oxidized form of coenzyme, NAD+ binding Active site Reaction 2. Enzyme II reacts with electron acceptor and reduced form of coenzyme, NADH binding Active site Enzyme II Enzyme I NAD+ Electron donor NADH Electron acceptor Enzyme substrate complex NADH Electron donor oxidized NAD+ Electron acceptor reduced
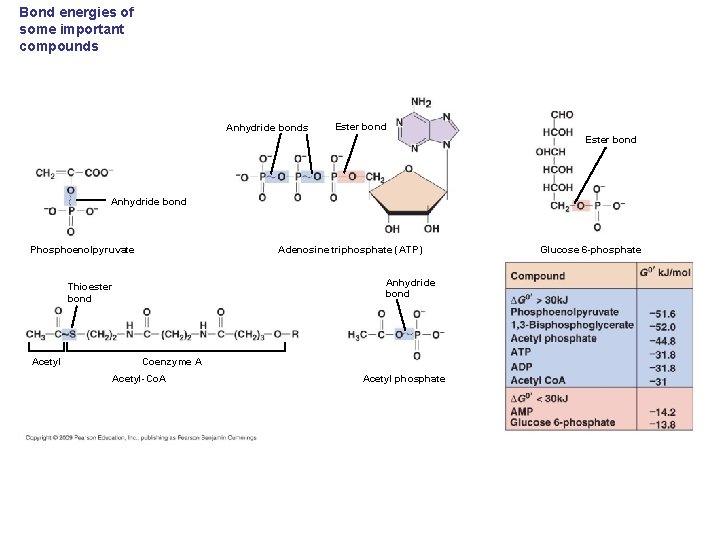
Bond energies of some important compounds Anhydride bonds Ester bond Anhydride bond Adenosine triphosphate (ATP) Phosphoenolpyruvate Anhydride bond Thioester bond Acetyl Coenzyme A Acetyl-Co. A Acetyl phosphate Glucose 6 -phosphate
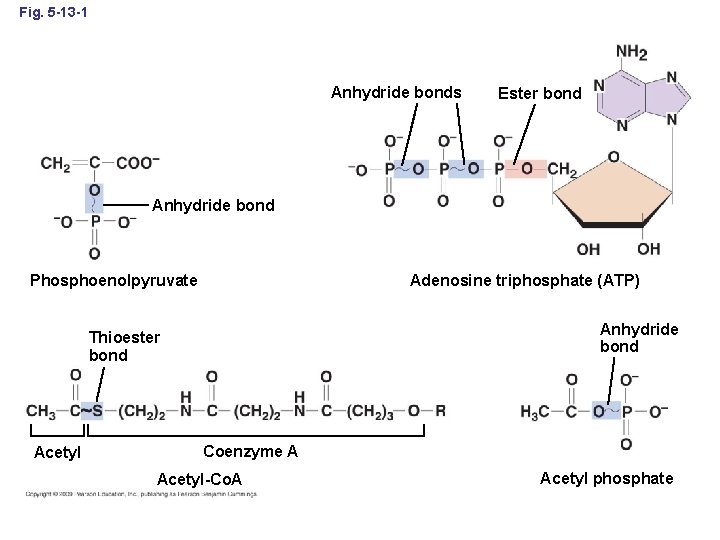
Fig. 5 -13 -1 Anhydride bonds Ester bond Anhydride bond Adenosine triphosphate (ATP) Phosphoenolpyruvate Anhydride bond Thioester bond Acetyl Coenzyme A Acetyl-Co. A Acetyl phosphate
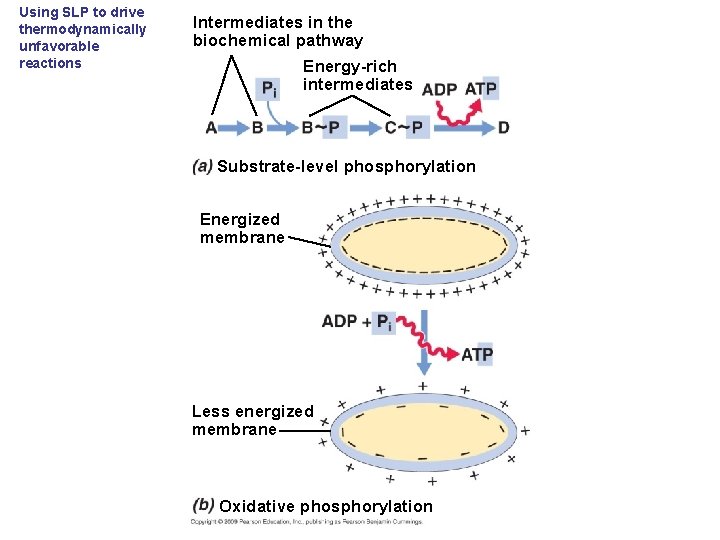
Using SLP to drive thermodynamically unfavorable reactions Intermediates in the biochemical pathway Energy-rich intermediates Substrate-level phosphorylation Energized membrane Less energized membrane Oxidative phosphorylation
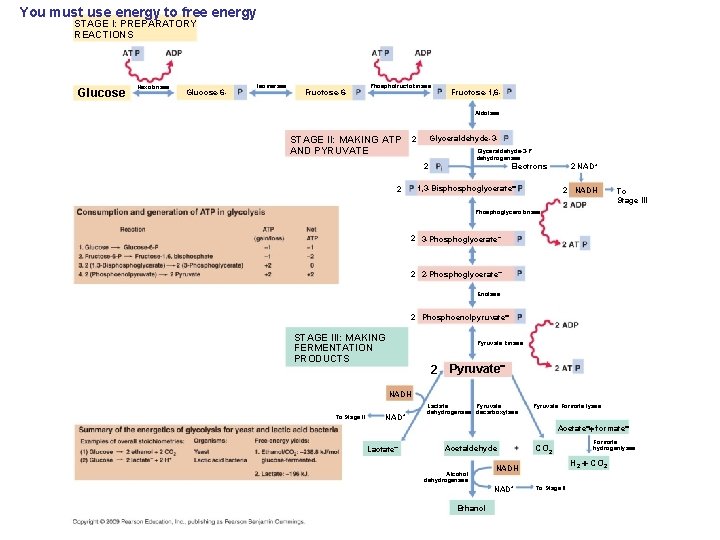
You must use energy to free energy STAGE I: PREPARATORY REACTIONS Glucose Hexokinase Glucose-6 - Isomerase Fructose-6 - Phosphofructokinase Fructose-1, 6 Aldolase STAGE II: MAKING ATP AND PYRUVATE Glyceraldehyde-3 - 2 Glyceraldehyde-3 -P dehydrogenase 2 Electrons 1, 3 -Bisphoglycerate 2 2 NAD+ 2 NADH To Stage III Phosphoglycerokinase 2 3 -Phosphoglycerate 2 2 -Phosphoglycerate Enolase 2 Phosphoenolpyruvate STAGE III: MAKING FERMENTATION PRODUCTS Pyruvate kinase 2 Pyruvate NADH To Stage II NAD+ Lactate Pyruvate dehydrogenase decarboxylase Pyruvate: Formate lyase Acetate formate Lactate Acetaldehyde Alcohol dehydrogenase Formate hydrogenlyase H 2 CO 2 NADH NAD+ Ethanol CO 2 To Stage II
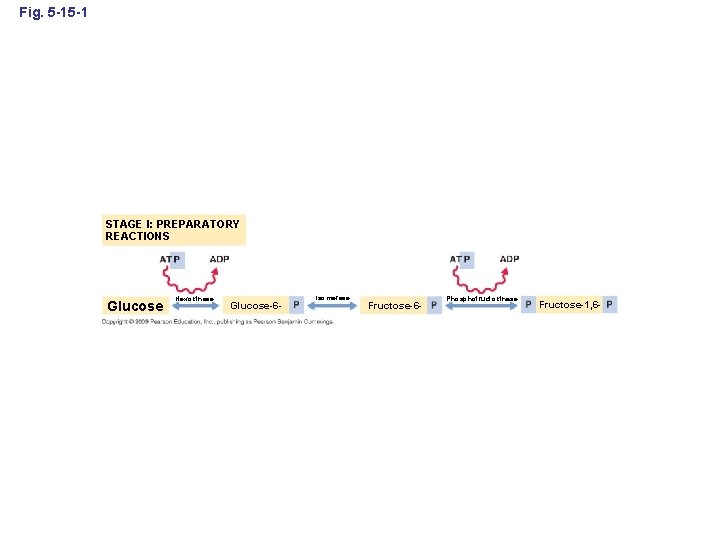
Fig. 5 -15 -1 STAGE I: PREPARATORY REACTIONS Glucose Hexokinase Glucose-6 - Isomerase Fructose-6 - Phosphofructokinase Fructose-1, 6 -
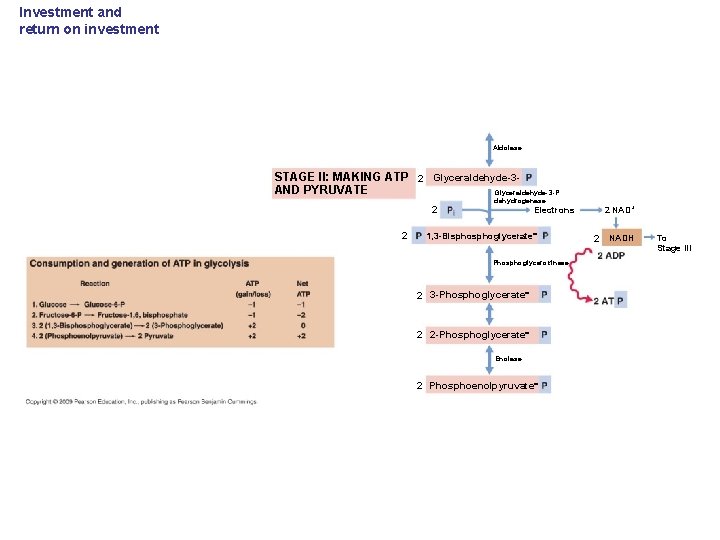
Investment and return on investment Aldolase STAGE II: MAKING ATP 2 Glyceraldehyde-3 AND PYRUVATE Glyceraldehyde-3 -P 2 2 dehydrogenase Electrons 1, 3 -Bisphoglycerate Phosphoglycerokinase 2 3 -Phosphoglycerate 2 2 -Phosphoglycerate Enolase 2 Phosphoenolpyruvate 2 NAD+ 2 NADH To Stage III
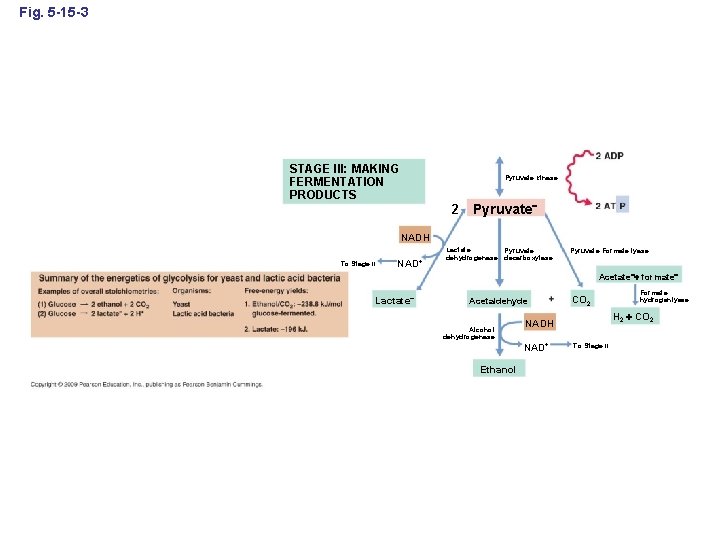
Fig. 5 -15 -3 STAGE III: MAKING FERMENTATION PRODUCTS Pyruvate kinase 2 Pyruvate NADH To Stage II NAD+ Lactate Pyruvate dehydrogenase decarboxylase Pyruvate: Formate lyase Acetate formate Lactate Acetaldehyde Alcohol dehydrogenase Formate hydrogenlyase H 2 CO 2 NADH NAD+ Ethanol CO 2 To Stage II
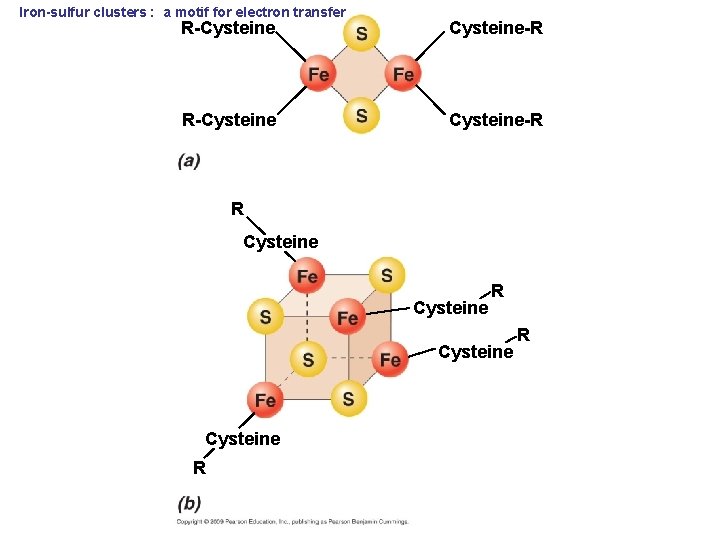
Iron-sulfur clusters : a motif for electron transfer R-Cysteine-R R Cysteine R R
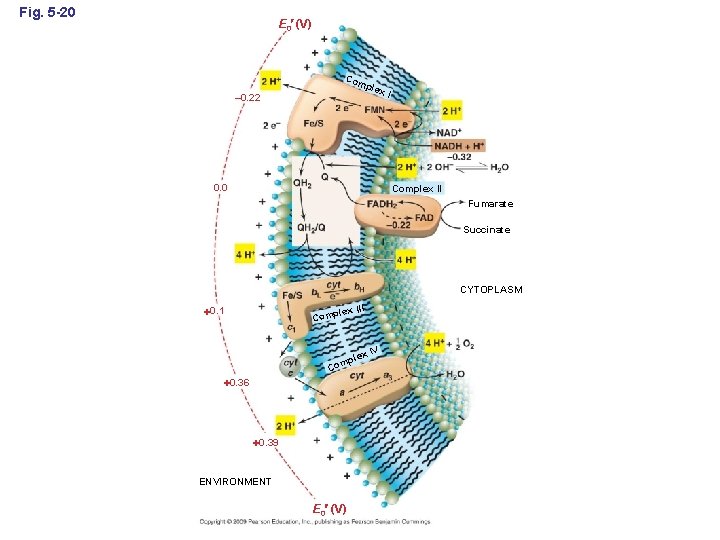
Fig. 5 -20 E 0 (V) Com plex – 0. 22 0. 0 I Complex II Fumarate Succinate CYTOPLASM lex III 0. 1 Comp x IV ple Com 0. 36 0. 39 ENVIRONMENT E 0 (V)
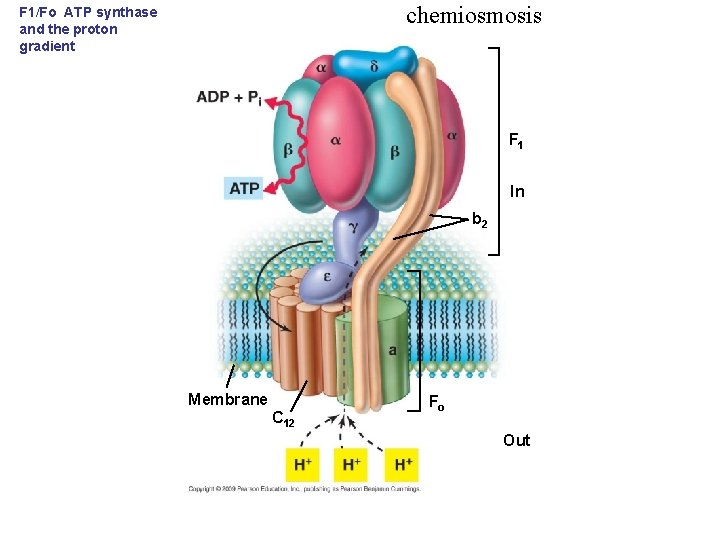
chemiosmosis F 1/Fo ATP synthase and the proton gradient F 1 In b 2 Membrane C 12 Fo Out
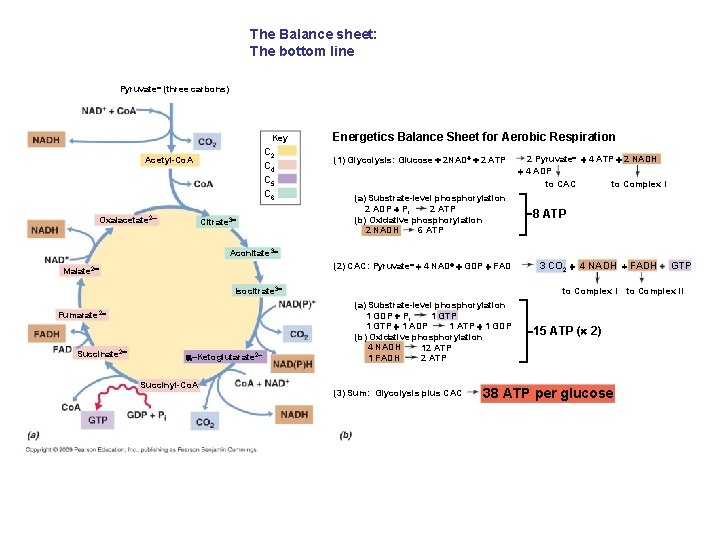
The Balance sheet: The bottom line Pyruvate (three carbons) Key C 2 C 4 C 5 C 6 Acetyl-Co. A Oxalacetate 2 Citrate 3 Energetics Balance Sheet for Aerobic Respiration (1) Glycolysis: Glucose 2 NAD 2 ATP (a) Substrate-level phosphorylation 2 ADP Pi 2 ATP (b) Oxidative phosphorylation 2 NADH 6 ATP 2 Pyruvate 4 ATP 2 NADH 4 ADP to CAC to Complex I 8 ATP Aconitate 3 (2) CAC: Pyruvate 4 NAD GDP FAD Malate 2 Isocitrate 3 Fumarate 2 Succinate 2 –Ketoglutarate 2 Succinyl-Co. A 3 CO 2 4 NADH FADH to Complex I (a) Substrate-level phosphorylation 1 GDP Pi 1 GTP 1 ATP 1 GDP 1 GTP 1 ADP (b) Oxidative phosphorylation 4 NADH 12 ATP 1 FADH 2 ATP (3) Sum: Glycolysis plus CAC 15 ATP ( 2) 38 ATP per glucose GTP to Complex II
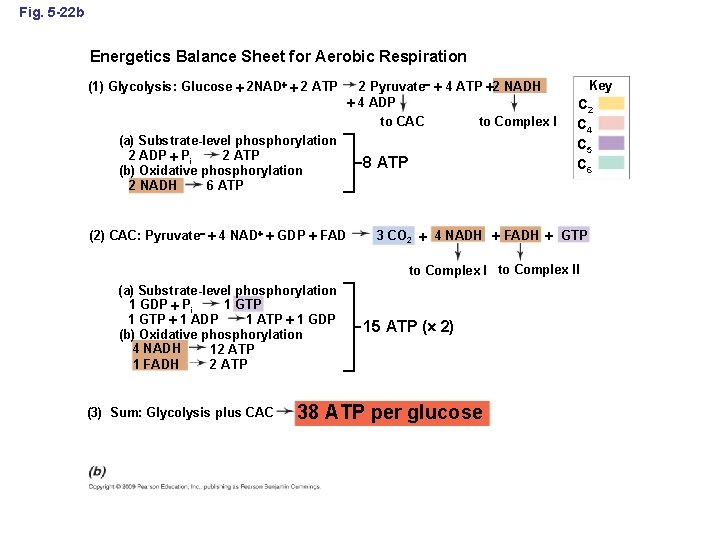
Fig. 5 -22 b Energetics Balance Sheet for Aerobic Respiration (1) Glycolysis: Glucose 2 NAD 2 ATP (a) Substrate-level phosphorylation 2 ATP 2 ADP Pi (b) Oxidative phosphorylation 6 ATP 2 NADH (2) CAC: Pyruvate 4 NAD GDP FAD 2 Pyruvate 4 ATP 2 NADH 4 ADP to CAC to Complex I 8 ATP Key C 2 C 4 C 5 C 6 3 CO 2 4 NADH FADH GTP to Complex II (a) Substrate-level phosphorylation 1 GTP 1 GDP Pi 1 GTP 1 ADP 1 ATP 1 GDP (b) Oxidative phosphorylation 4 NADH 12 ATP 1 FADH 2 ATP (3) Sum: Glycolysis plus CAC 15 ATP ( 2) 38 ATP per glucose
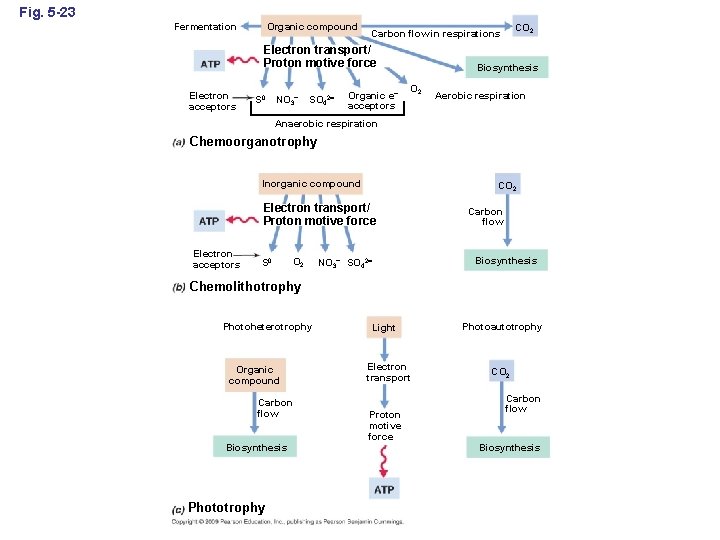
Fig. 5 -23 Fermentation Organic compound Electron transport/ Proton motive force Electron acceptors S 0 NO 3– SO 42 CO 2 Carbon flow in respirations Organic e– acceptors Biosynthesis O 2 Aerobic respiration Anaerobic respiration Chemoorganotrophy Inorganic compound CO 2 Electron transport/ Proton motive force Electron acceptors S 0 O 2 Carbon flow Biosynthesis NO 3– SO 42 Chemolithotrophy Photoheterotrophy Organic compound Carbon flow Biosynthesis Phototrophy Light Electron transport Proton motive force Photoautotrophy CO 2 Carbon flow Biosynthesis
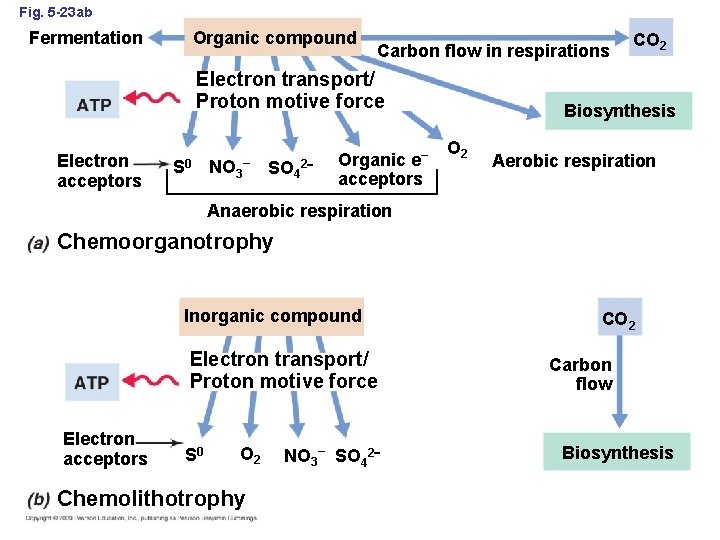
Fig. 5 -23 ab Organic compound Fermentation Carbon flow in respirations Electron transport/ Proton motive force Electron acceptors S 0 NO 3 – SO 42 Organic e– acceptors CO 2 Biosynthesis O 2 Aerobic respiration Anaerobic respiration Chemoorganotrophy Inorganic compound Electron transport/ Proton motive force Electron acceptors S 0 O 2 Chemolithotrophy NO 3– SO 42 CO 2 Carbon flow Biosynthesis
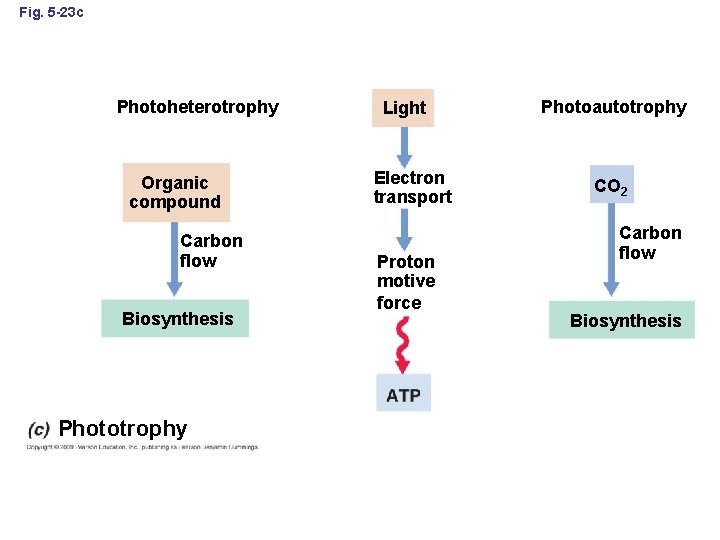
Fig. 5 -23 c Photoheterotrophy Organic compound Carbon flow Biosynthesis Phototrophy Light Electron transport Proton motive force Photoautotrophy CO 2 Carbon flow Biosynthesis
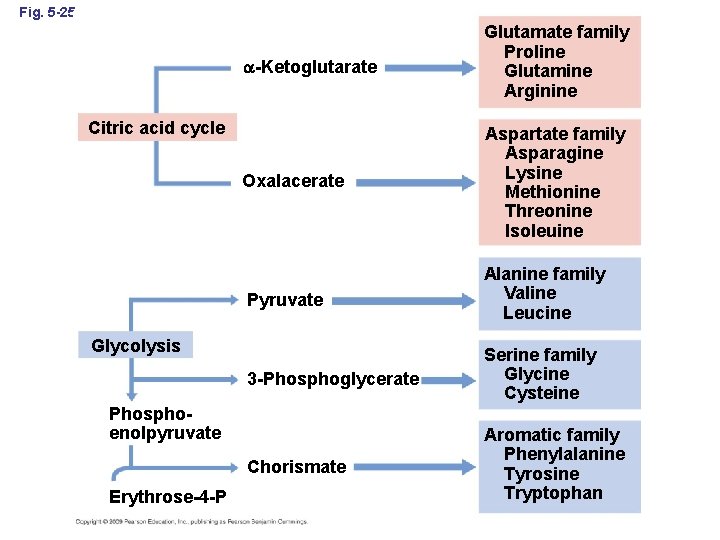
Fig. 5 -25 -Ketoglutarate Glutamate family Proline Glutamine Arginine Oxalacerate Aspartate family Asparagine Lysine Methionine Threonine Isoleuine Pyruvate Alanine family Valine Leucine 3 -Phosphoglycerate Serine family Glycine Cysteine Chorismate Aromatic family Phenylalanine Tyrosine Tryptophan Citric acid cycle Glycolysis Phosphoenolpyruvate Erythrose-4 -P
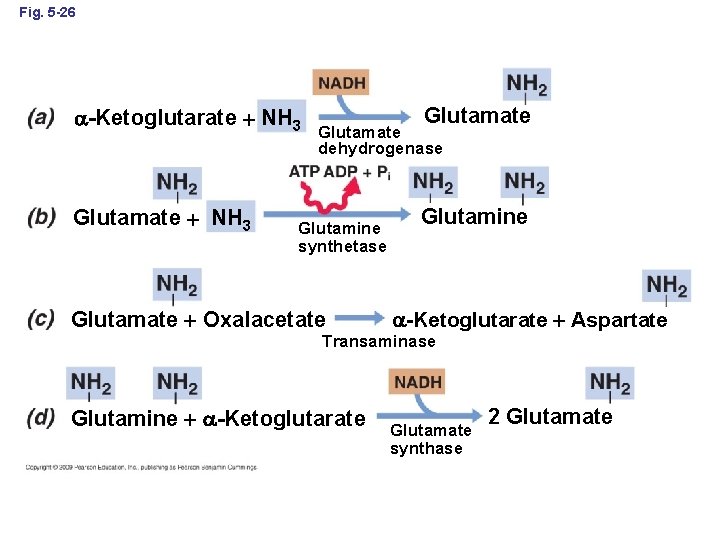
Fig. 5 -26 -Ketoglutarate NH 3 Glutamate dehydrogenase Glutamine synthetase Glutamate Oxalacetate Glutamine -Ketoglutarate Aspartate Transaminase Glutamine -Ketoglutarate Glutamate synthase 2 Glutamate
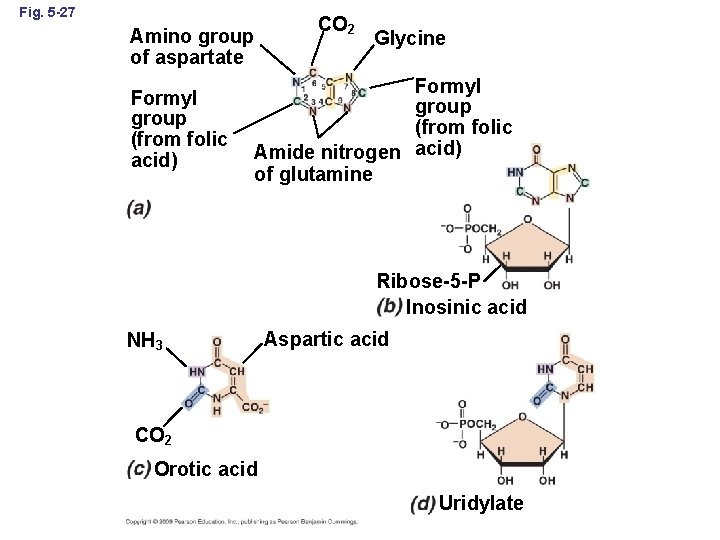
Fig. 5 -27 Amino group of aspartate Formyl group (from folic acid) CO 2 Glycine Formyl group (from folic Amide nitrogen acid) of glutamine Ribose-5 -P Inosinic acid NH 3 Aspartic acid CO 2 Orotic acid Uridylate
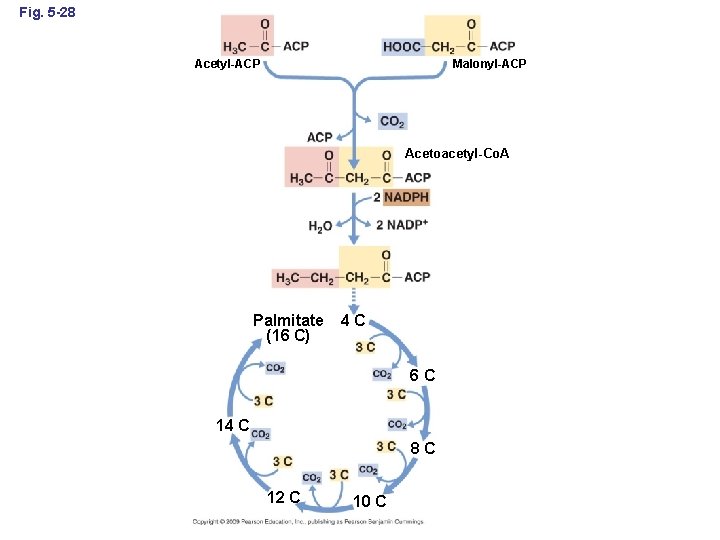
Fig. 5 -28 Acetyl-ACP Malonyl-ACP Acetoacetyl-Co. A Palmitate (16 C) 4 C 6 C 14 C 8 C 12 C 10 C
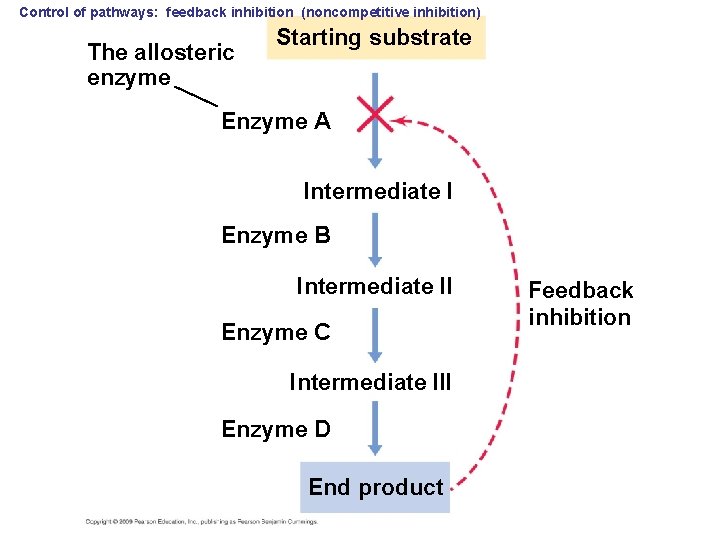
Control of pathways: feedback inhibition (noncompetitive inhibition) The allosteric enzyme Starting substrate Enzyme A Intermediate I Enzyme B Intermediate II Enzyme C Intermediate III Enzyme D End product Feedback inhibition
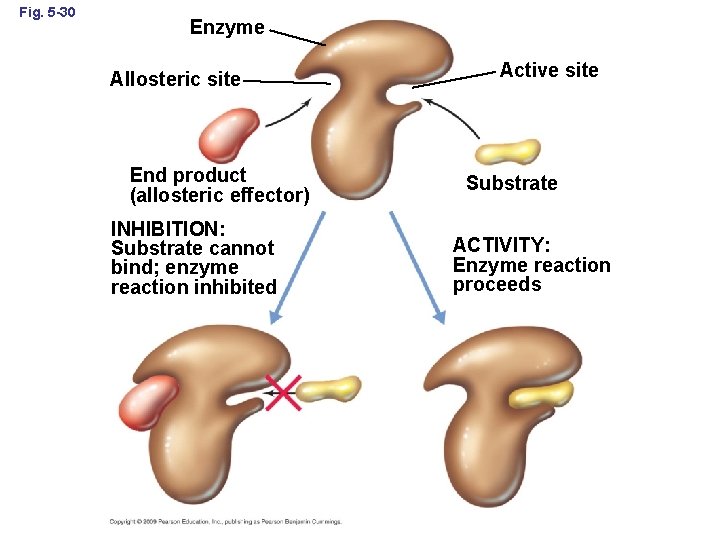
Fig. 5 -30 Enzyme Allosteric site End product (allosteric effector) INHIBITION: Substrate cannot bind; enzyme reaction inhibited Active site Substrate ACTIVITY: Enzyme reaction proceeds
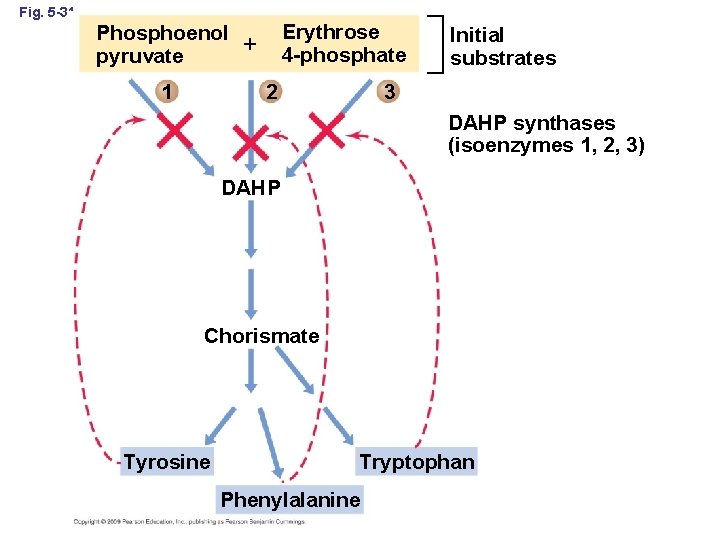
Fig. 5 -31 Erythrose 4 -phosphate Phosphoenol pyruvate 1 Initial substrates 3 2 DAHP synthases (isoenzymes 1, 2, 3) DAHP Chorismate Tyrosine Tryptophan Phenylalanine
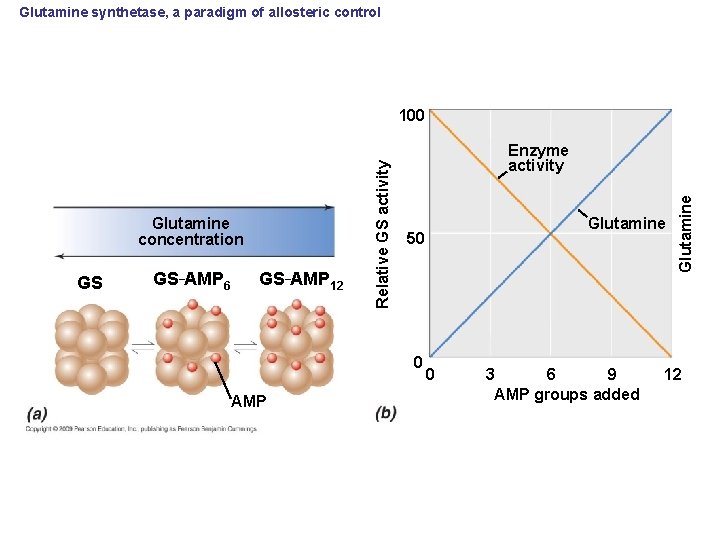
Glutamine synthetase, a paradigm of allosteric control GS GS–AMP 6 GS–AMP 12 Enzyme activity 0 AMP Glutamine 50 0 3 6 9 AMP groups added Glutamine concentration Relative GS activity 100 12
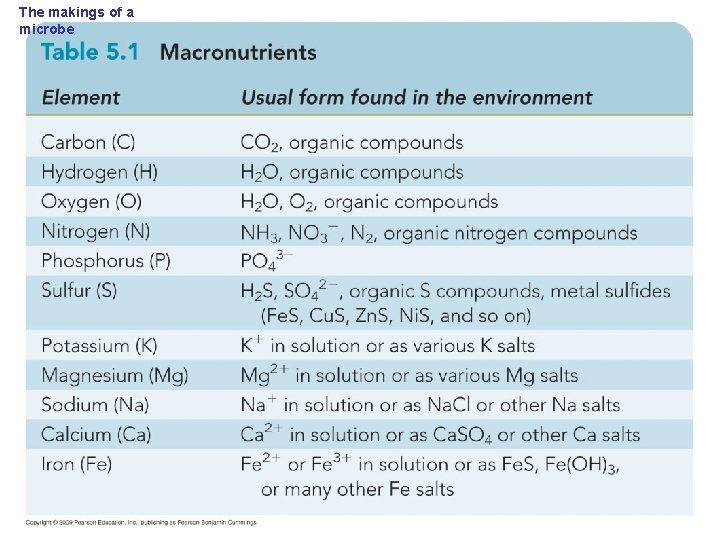
The makings of a microbe
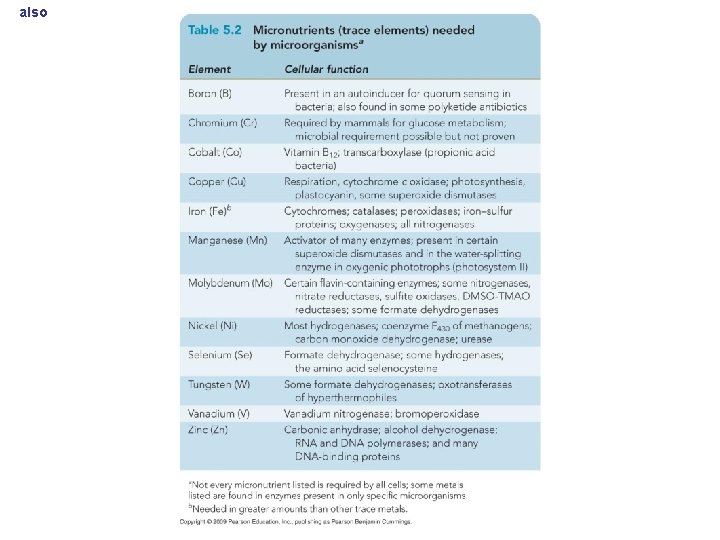
also
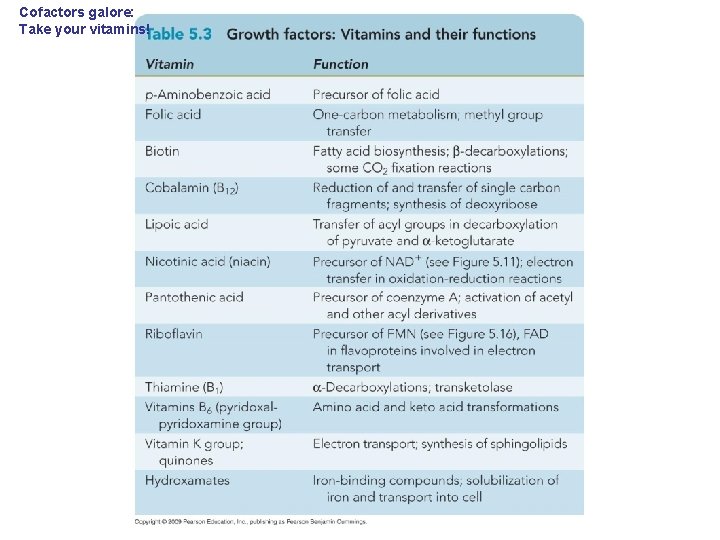
Cofactors galore: Take your vitamins!
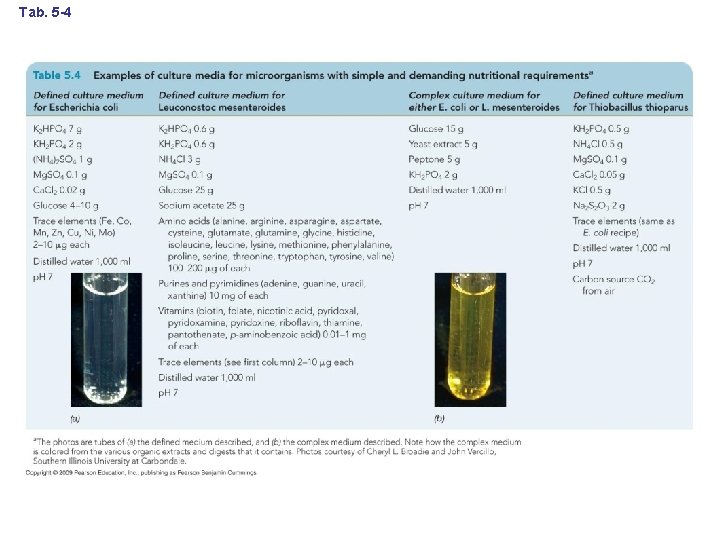
Tab. 5 -4
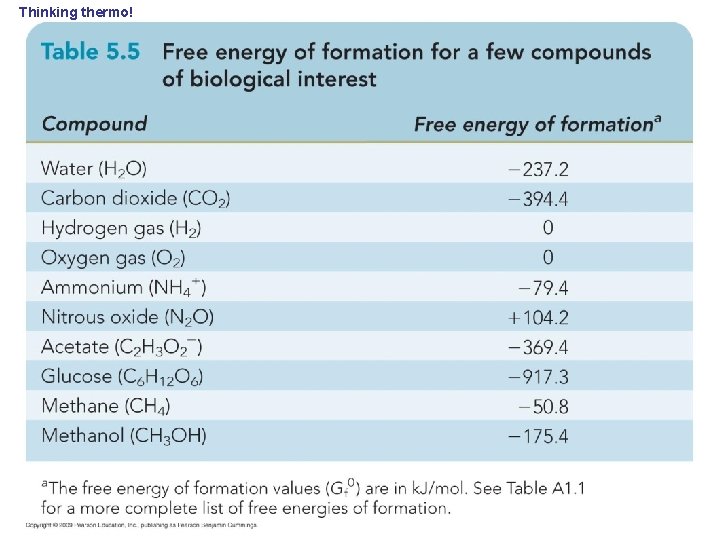
Thinking thermo!
- Slides: 40