Microbiology A Systems Approach Chapter 9 Microbial Genetics

Microbiology: A Systems Approach Chapter 9: Microbial Genetics
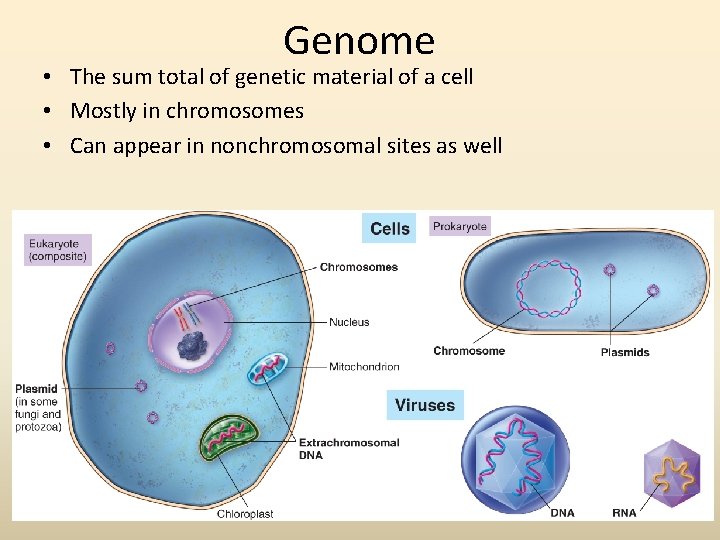
Genome • The sum total of genetic material of a cell • Mostly in chromosomes • Can appear in nonchromosomal sites as well
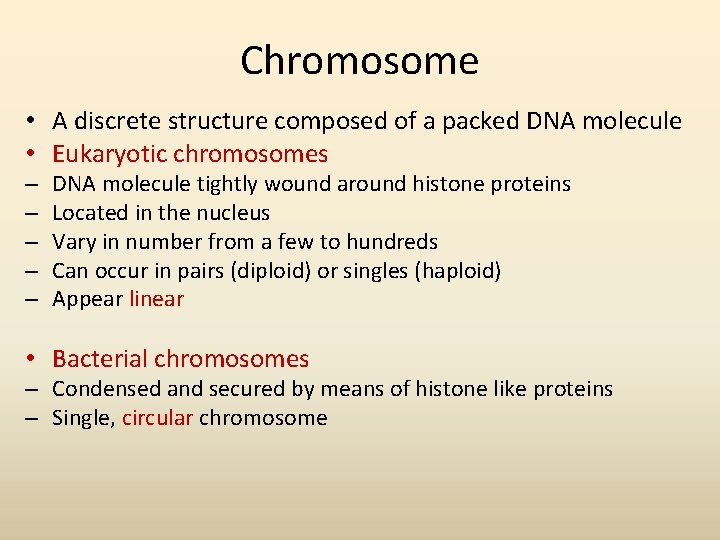
Chromosome • A discrete structure composed of a packed DNA molecule • Eukaryotic chromosomes – – – DNA molecule tightly wound around histone proteins Located in the nucleus Vary in number from a few to hundreds Can occur in pairs (diploid) or singles (haploid) Appear linear • Bacterial chromosomes – Condensed and secured by means of histone like proteins – Single, circular chromosome
Gene • A segment of DNA that codes for RNA and protein • Genotype- genetic makeup • Phenotype- expression of the genotype (traits) The Size and Packaging of Genomes • Vary greatly in size – Viruses- 4 -100 genes – E. coli- 4, 000 genes – Human cell- 25, 000 to 40, 000 genes • The stretched-out DNA can be 1, 000 times or more longer than the cell
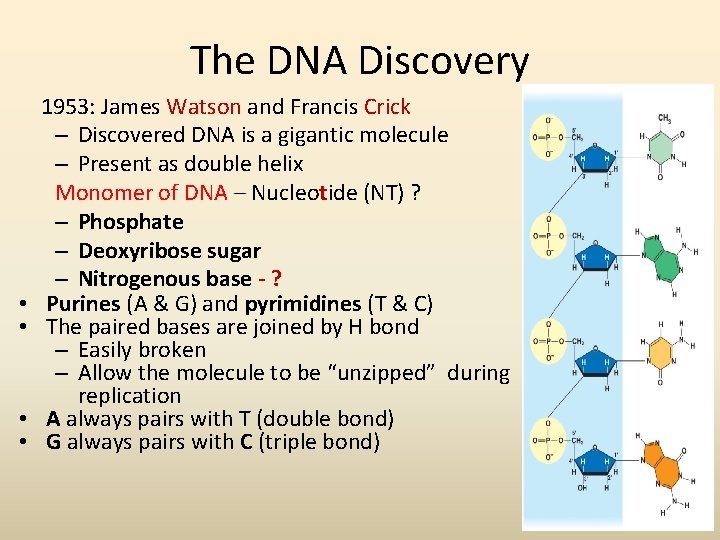
The DNA Discovery • • 1953: James Watson and Francis Crick – Discovered DNA is a gigantic molecule – Present as double helix Monomer of DNA – Nucleotide (NT) ? – Phosphate – Deoxyribose sugar – Nitrogenous base - ? Purines (A & G) and pyrimidines (T & C) The paired bases are joined by H bond – Easily broken – Allow the molecule to be “unzipped” during replication A always pairs with T (double bond) G always pairs with C (triple bond)
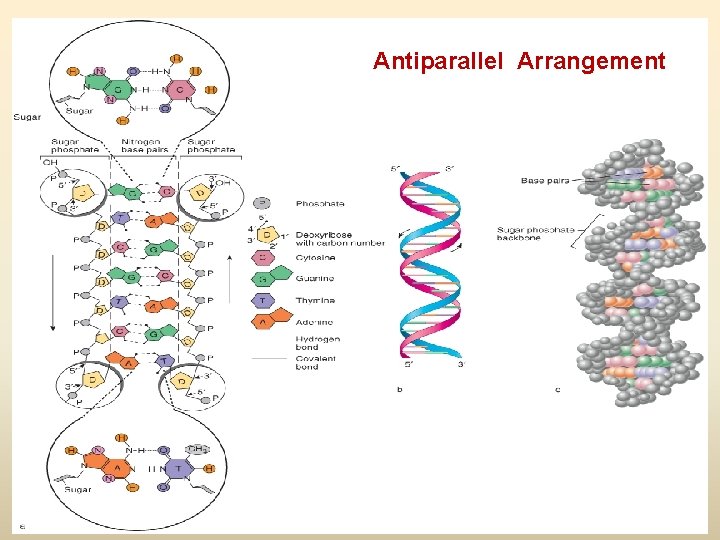
Antiparallel Arrangement
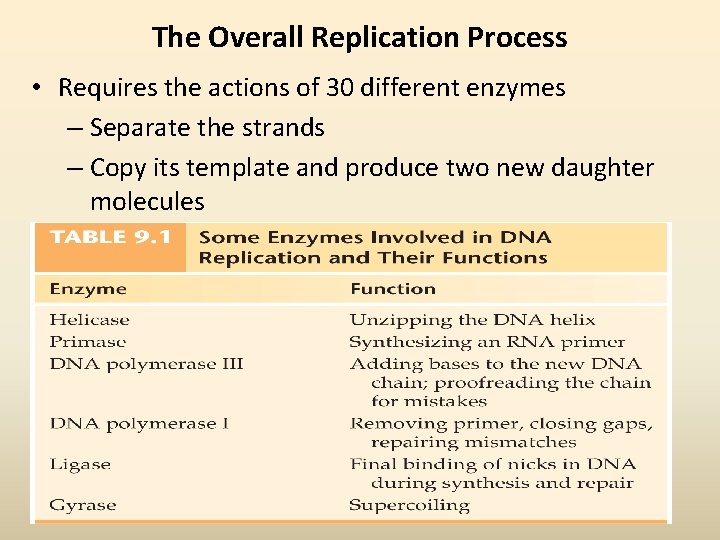
The Overall Replication Process • Requires the actions of 30 different enzymes – Separate the strands – Copy its template and produce two new daughter molecules
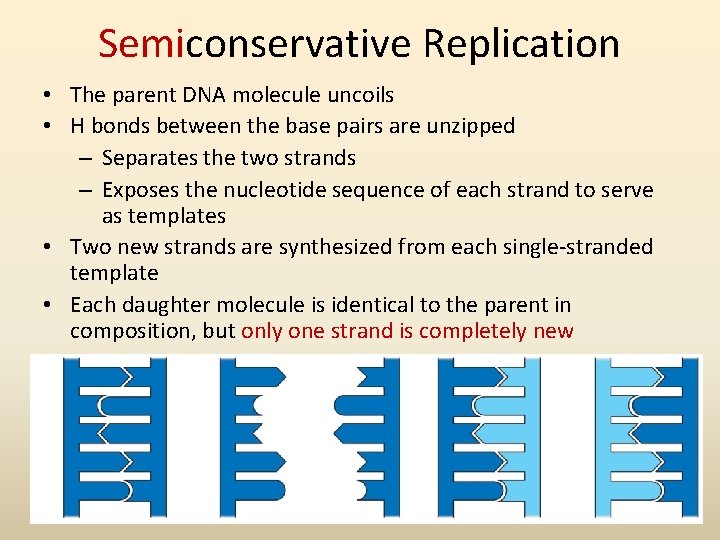
Semiconservative Replication • The parent DNA molecule uncoils • H bonds between the base pairs are unzipped – Separates the two strands – Exposes the nucleotide sequence of each strand to serve as templates • Two new strands are synthesized from each single-stranded template • Each daughter molecule is identical to the parent in composition, but only one strand is completely new
Process of Replication • Origin of replication – Short sequence, Rich in A and T – Less energy is required to separate the two strands • Helicases bind to the DNA at the origin – Untwist the helix – Break the hydrogen bonds – Results in two separate strands • DNA Polymerase III - Synthesizes a new strand using template strand - DNA Poly III add NTs to an already existing chain-short primer (RNA primer made by Primase as DNA poly III can not work from scratch) - DNA polymerase III can only add NTs in one direction - A new strand is always synthesized from 5’ to 3’ direction
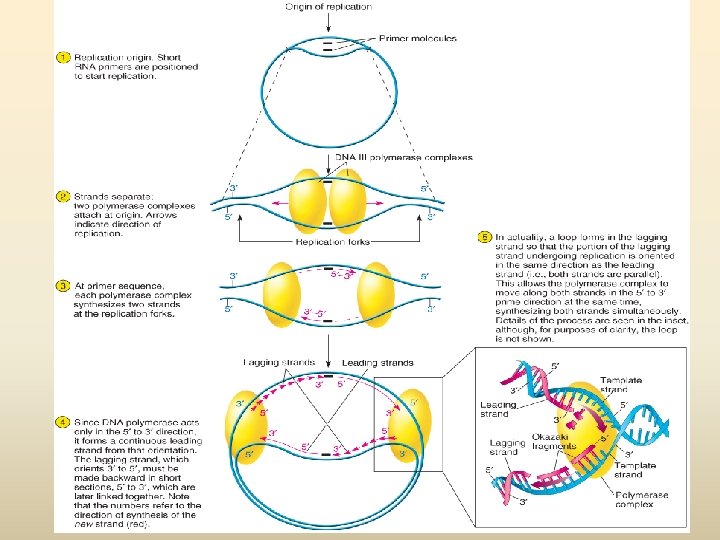
Elongation and Termination of the Daughter Molecules • DNA Poly I removes RNA primers and replaces them with DNA • When the forks come full circle and meet, ligases move along the lagging strand – link the fragments result in synthesis and separation of the two daughter molecules – Occasionally an incorrect base is added to the growing chain • Most are corrected; If not corrected, result in mutations • DNA Poly III can detect incorrect, unmatching bases, excise them, and replace them with the correct base • DNA Poly I can also proofread and repair
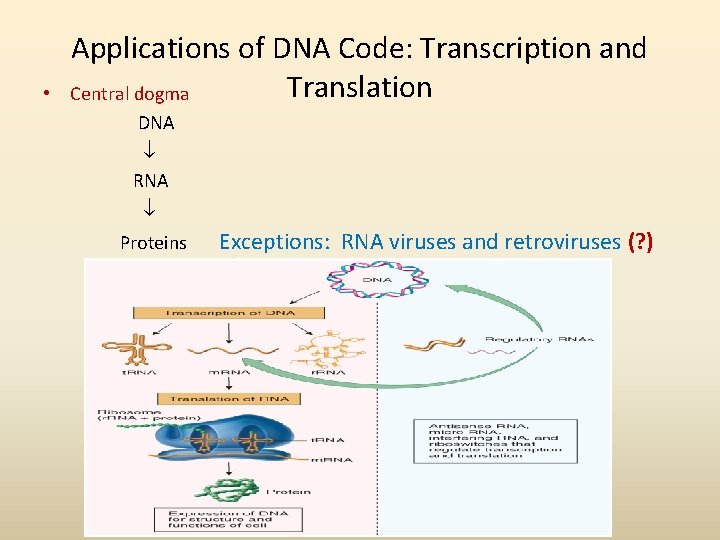
• Applications of DNA Code: Transcription and Translation Central dogma DNA RNA Proteins Exceptions: RNA viruses and retroviruses (? )
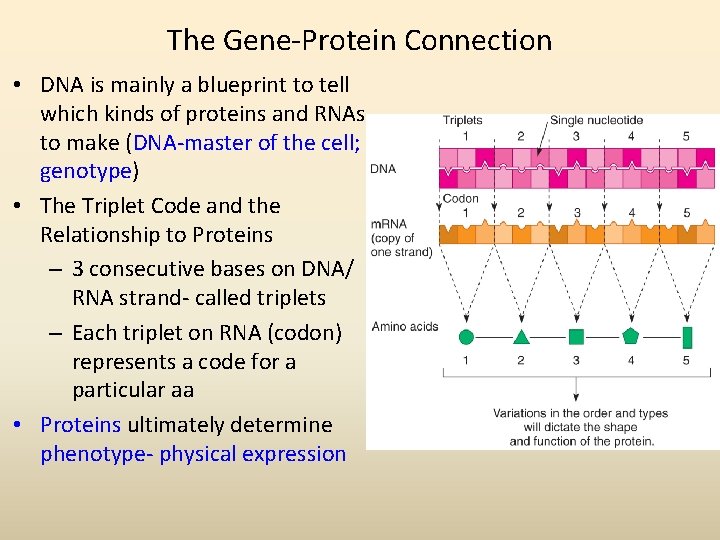
The Gene-Protein Connection • DNA is mainly a blueprint to tell which kinds of proteins and RNAs to make (DNA-master of the cell; genotype) • The Triplet Code and the Relationship to Proteins – 3 consecutive bases on DNA/ RNA strand- called triplets – Each triplet on RNA (codon) represents a code for a particular aa • Proteins ultimately determine phenotype- physical expression
Transcription and Translation • A number of components participate including: – m. RNA, t. RNA, ribosomes (r. RNA) – regulatory RNAs, several enzymes and raw materials • RNAs- differs from DNA • Single stranded molecule • Contains uracil instead of thymine • The sugar is ribose (Name sugar in DNA; is it pentose or hexose? ) – Only m. RNA is translated into a protein molecule
Transcription produces genetic messages in the form of RNA Stages of transcription – Initiation: RNA Polymerase (Poly) binds to a promoter, where the helix unwinds and transcription starts – Elongation: RNA NTs are added to the chain – Termination: RNA Poly reaches a terminator sequence and detaches from the template – 3 different RNAs: m. RNA, r. RNA and t. RNA Copyright © 2009 Pearson Education, Inc.
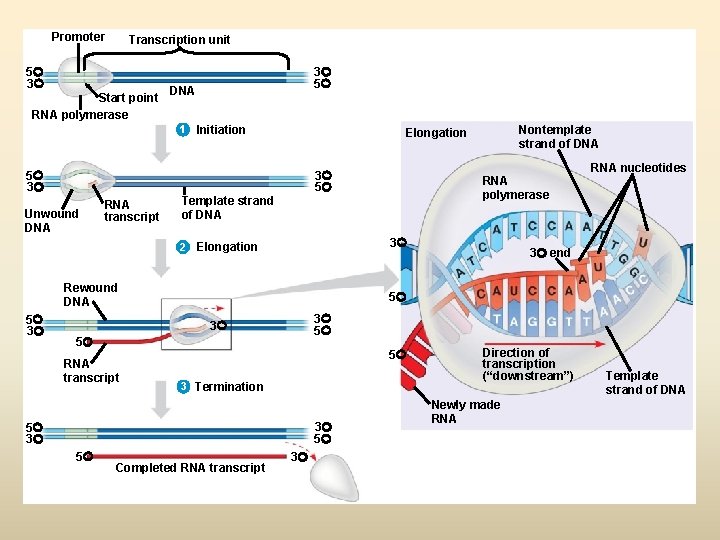
Promoter Transcription unit 5 3 Start point RNA polymerase 3 5 DNA 1 Initiation 5 3 3 5 Unwound DNA RNA transcript 3 Rewound DNA 3 end 3 5 5 5 3 Termination 3 5 5 3 5 RNA nucleotides 5 3 RNA transcript RNA polymerase Template strand of DNA 2 Elongation 5 3 Nontemplate strand of DNA Elongation Completed RNA transcript 3 Direction of transcription (“downstream”) Newly made RNA Template strand of DNA
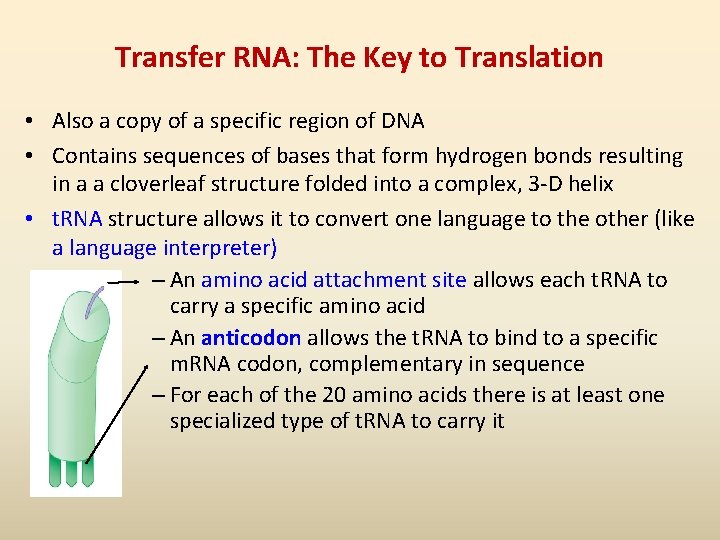
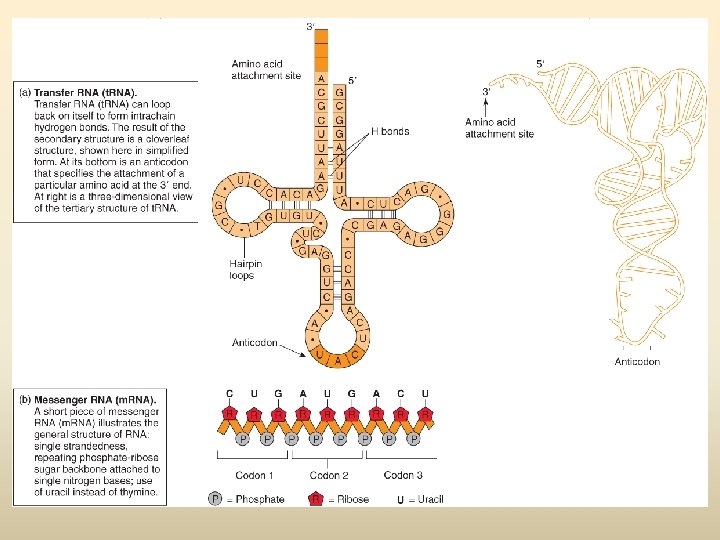
Transfer RNA: The Key to Translation • Also a copy of a specific region of DNA • Contains sequences of bases that form hydrogen bonds resulting in a a cloverleaf structure folded into a complex, 3 -D helix • t. RNA structure allows it to convert one language to the other (like a language interpreter) – An amino acid attachment site allows each t. RNA to carry a specific amino acid – An anticodon allows the t. RNA to bind to a specific m. RNA codon, complementary in sequence – For each of the 20 amino acids there is at least one specialized type of t. RNA to carry it
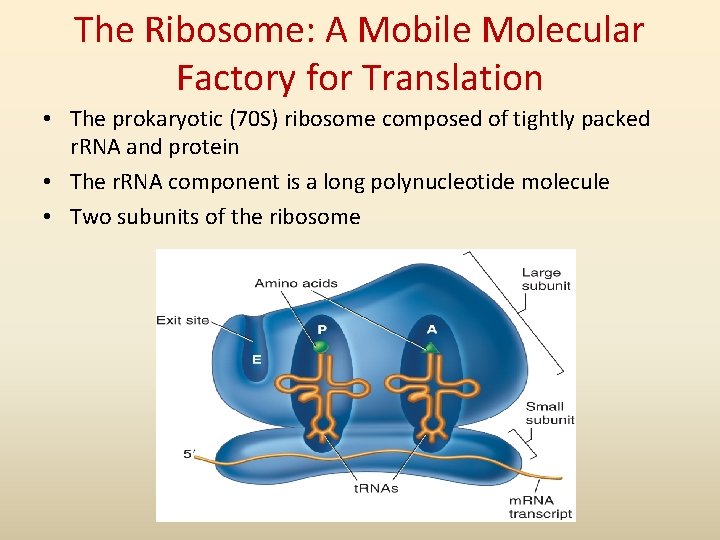
The Ribosome: A Mobile Molecular Factory for Translation • The prokaryotic (70 S) ribosome composed of tightly packed r. RNA and protein • The r. RNA component is a long polynucleotide molecule • Two subunits of the ribosome
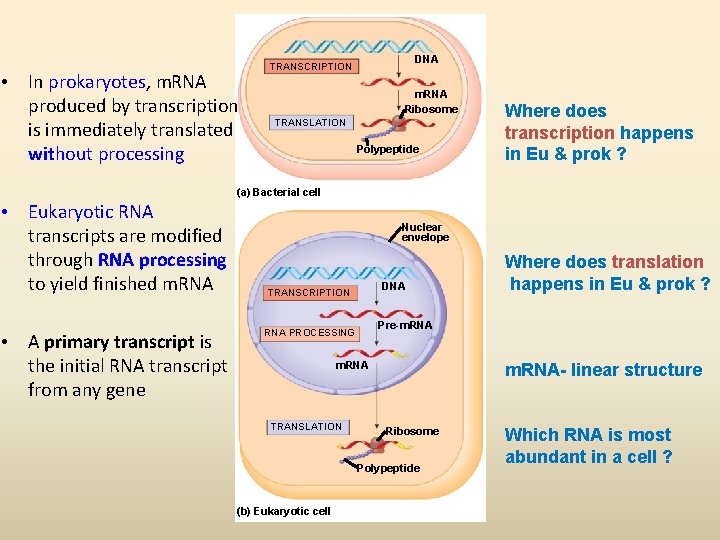
• In prokaryotes, m. RNA produced by transcription is immediately translated without processing DNA TRANSCRIPTION m. RNA Ribosome TRANSLATION Polypeptide Where does transcription happens in Eu & prok ? (a) Bacterial cell • Eukaryotic RNA transcripts are modified through RNA processing to yield finished m. RNA • A primary transcript is the initial RNA transcript from any gene Nuclear envelope DNA TRANSCRIPTION Pre-m. RNA PROCESSING m. RNA TRANSLATION m. RNA- linear structure Ribosome Polypeptide (b) Eukaryotic cell Where does translation happens in Eu & prok ? Which RNA is most abundant in a cell ?
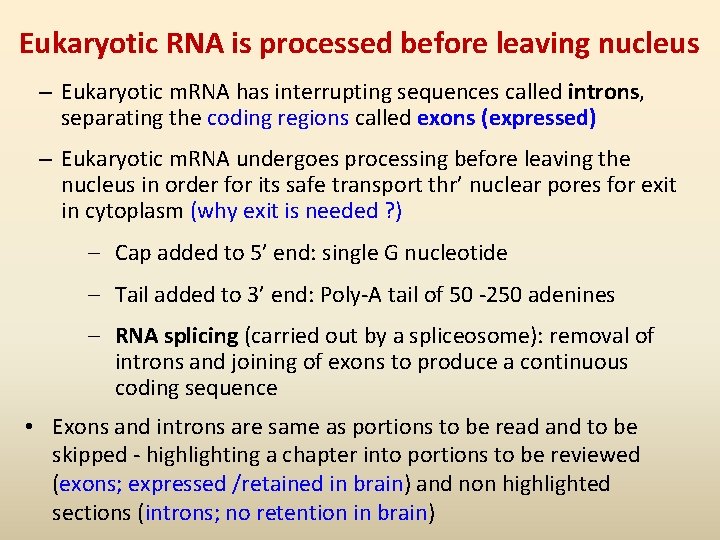
Eukaryotic RNA is processed before leaving nucleus – Eukaryotic m. RNA has interrupting sequences called introns, separating the coding regions called exons (expressed) – Eukaryotic m. RNA undergoes processing before leaving the nucleus in order for its safe transport thr’ nuclear pores for exit in cytoplasm (why exit is needed ? ) – Cap added to 5’ end: single G nucleotide – Tail added to 3’ end: Poly-A tail of 50 -250 adenines – RNA splicing (carried out by a spliceosome): removal of introns and joining of exons to produce a continuous coding sequence • Exons and introns are same as portions to be read and to be skipped - highlighting a chapter into portions to be reviewed (exons; expressed /retained in brain) and non highlighted sections (introns; no retention in brain)
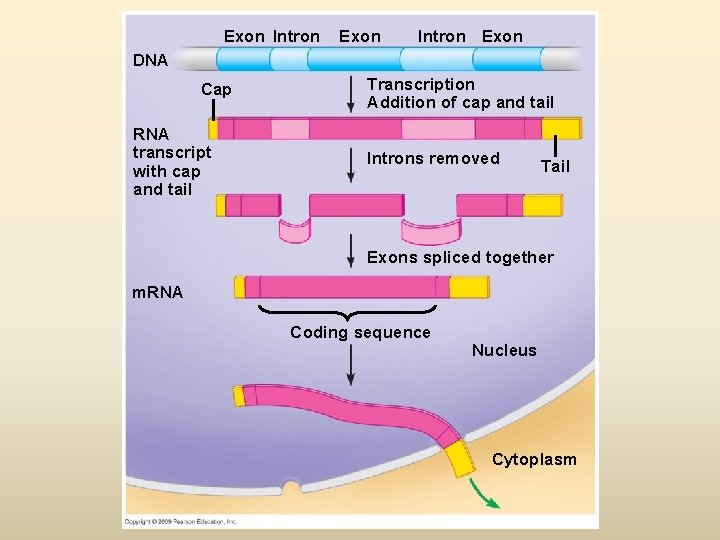
Exon Intron Exon DNA Cap RNA transcript with cap and tail Transcription Addition of cap and tail Introns removed Tail Exons spliced together m. RNA Coding sequence Nucleus Cytoplasm
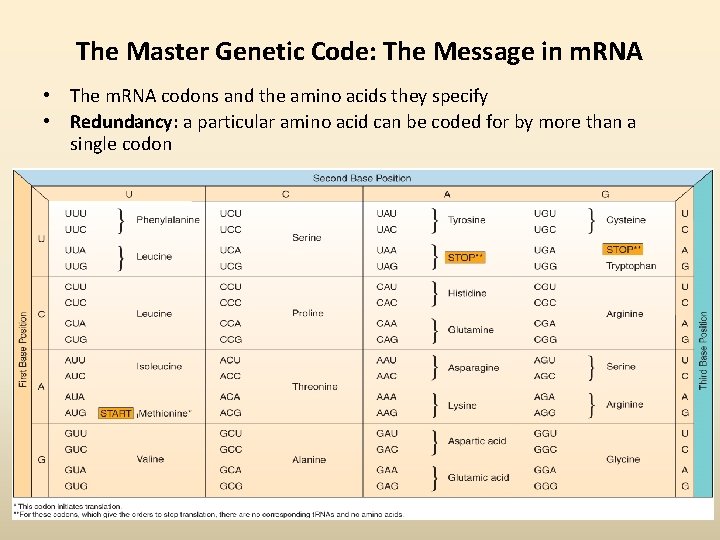
The Master Genetic Code: The Message in m. RNA • The m. RNA codons and the amino acids they specify • Redundancy: a particular amino acid can be coded for by more than a single codon
Translation: 2 nd Stage of Gene Expression • Ribosomes are the sites of translation • Prokaryotic ribosomes -70 s size (50 s and 30 s subunit) • Eukaryotic ribosomes- 80 s (60 s and 40 s subunit) • 3 stages: initiation, elongation, termination
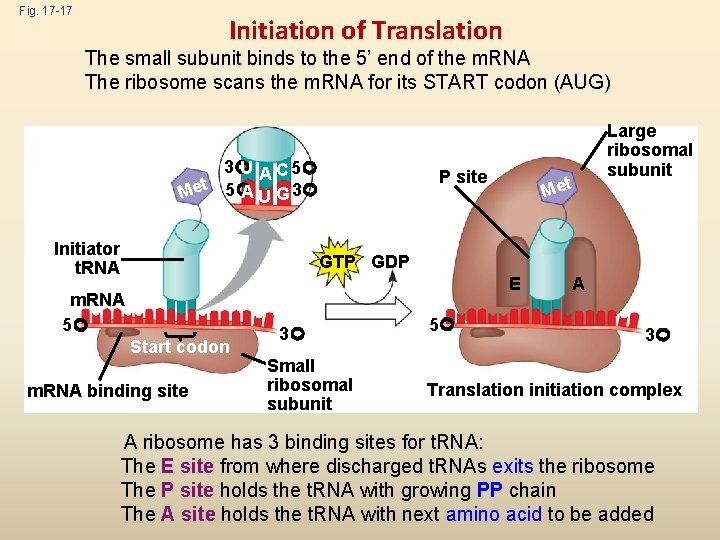
Fig. 17 -17 Initiation of Translation The small subunit binds to the 5’ end of the m. RNA The ribosome scans the m. RNA for its START codon (AUG) 3 U A C 5 A U G 3 Met 5 Initiator t. RNA P site Met Large ribosomal subunit GTP GDP E m. RNA 5 Start codon m. RNA binding site 3 Small ribosomal subunit 5 A 3 Translation initiation complex A ribosome has 3 binding sites for t. RNA: The E site from where discharged t. RNAs exits the ribosome The P site holds the t. RNA with growing PP chain The A site holds the t. RNA with next amino acid to be added
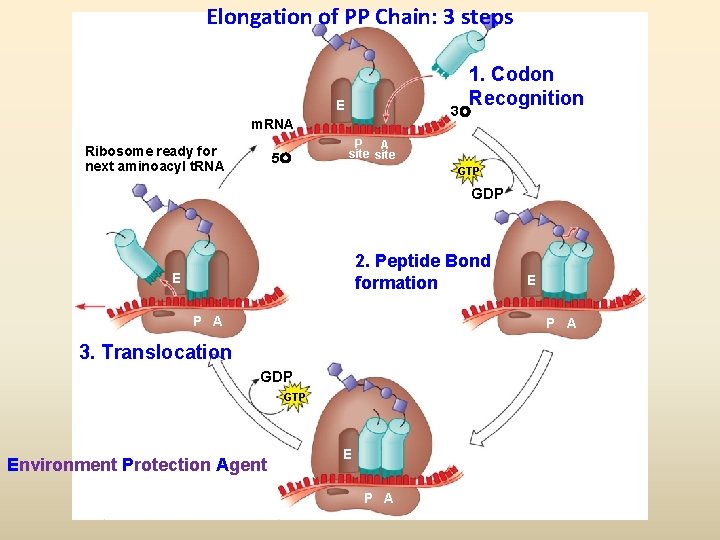
Elongation of PP Chain: 3 steps 1. Codon Recognition E 3 m. RNA Ribosome ready for next aminoacyl t. RNA 5 P A site GTP GDP 2. Peptide Bond formation E P A 3. Translocation GDP GTP Environment Protection Agent E P A
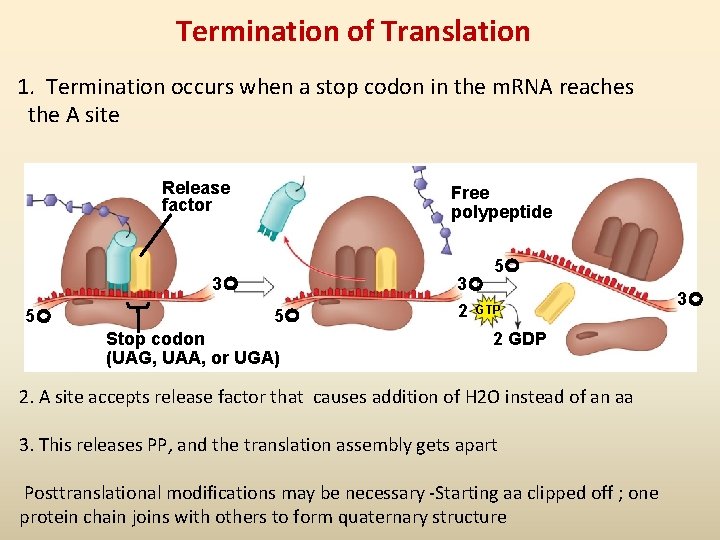
Termination of Translation 1. Termination occurs when a stop codon in the m. RNA reaches the A site Release factor Free polypeptide 5 3 5 5 Stop codon (UAG, UAA, or UGA) 3 2 GTP 2 GDP 2. A site accepts release factor that causes addition of H 2 O instead of an aa 3. This releases PP, and the translation assembly gets apart Posttranslational modifications may be necessary -Starting aa clipped off ; one protein chain joins with others to form quaternary structure 3
Genetic Regulation of Protein Synthesis and Metabolism • Control mechanisms ensure that genes are active only when their products are required – Enzymes are produced as they are needed – Prevents the waste of energy and materials • Prokaryotes organize collections of genes into operons – Coordinated set of genes regulated as a single unit – Either inducible or repressible • Inducible- the operon is turned ON by the substrate of the enzyme for which the structural genes code • Repressible- contain genes coding for anabolic enzymes; several genes in a series are turned OFF by the product synthesized by the enzyme
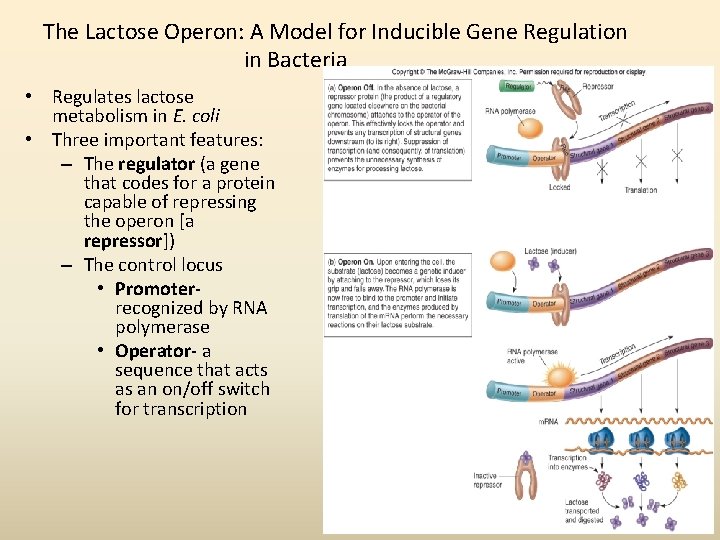
The Lactose Operon: A Model for Inducible Gene Regulation in Bacteria • Regulates lactose metabolism in E. coli • Three important features: – The regulator (a gene that codes for a protein capable of repressing the operon [a repressor]) – The control locus • Promoterrecognized by RNA polymerase • Operator- a sequence that acts as an on/off switch for transcription
Mutations: Changes in the Genetic Code • Mutation: An alteration in the N-base sequence of DNA; phenotypic changes happen due to changes in the genotype • Wild type: a natural, non mutated microbe • Mutant strain: when a microbe bears a mutation – Can show variance in morphology, nutritional characteristics, temp preference etc. e. g in culture of lactose + bacteria (which has capacity to utilize lactose), some bacteria have lost that capability (lactose -) Positive and Negative Effects of Mutations • Mutations are permanent and inheritable • Most are harmful but some provide adaptive advantages
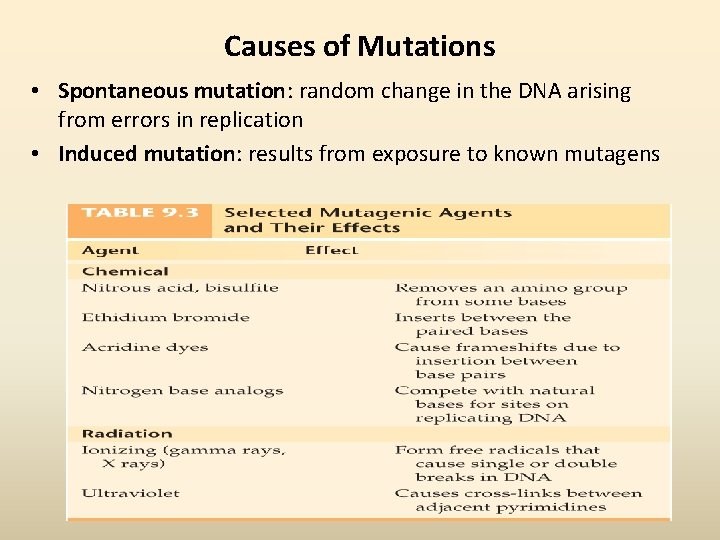
Causes of Mutations • Spontaneous mutation: random change in the DNA arising from errors in replication • Induced mutation: results from exposure to known mutagens
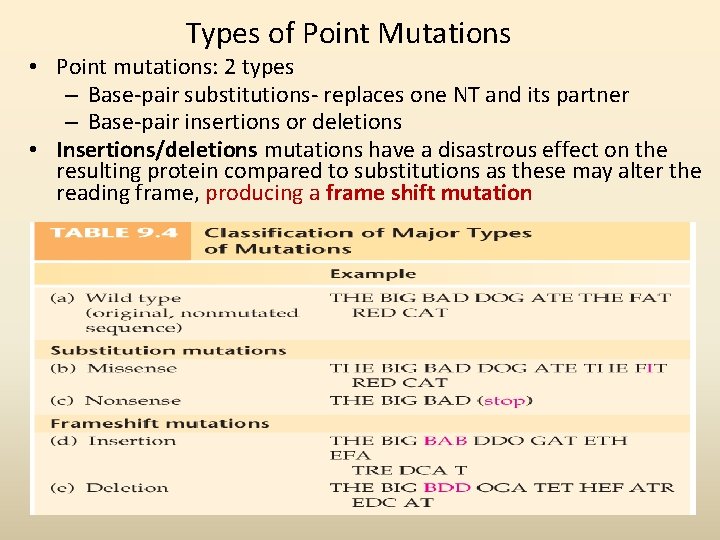
Types of Point Mutations • Point mutations: 2 types – Base-pair substitutions- replaces one NT and its partner – Base-pair insertions or deletions • Insertions/deletions mutations have a disastrous effect on the resulting protein compared to substitutions as these may alter the reading frame, producing a frame shift mutation
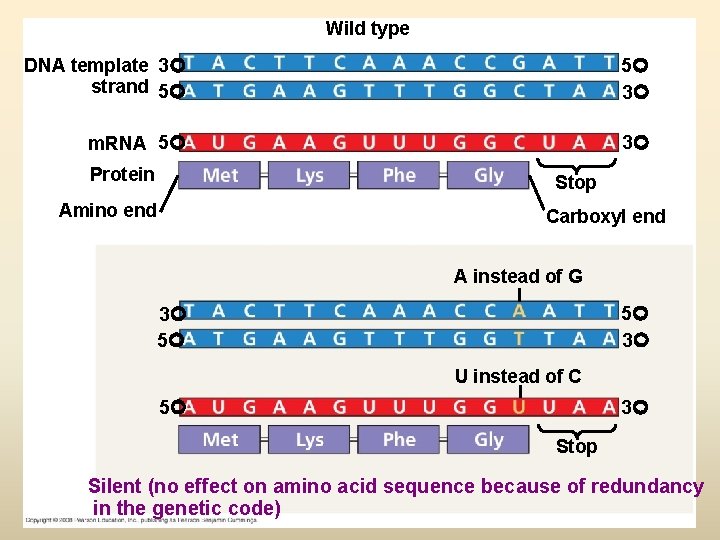
Wild type DNA template 3 strand 5 5 3 m. RNA 5 3 Protein Stop Amino end Carboxyl end A instead of G 5 3 3 5 U instead of C 5 3 Stop Silent (no effect on amino acid sequence because of redundancy in the genetic code)
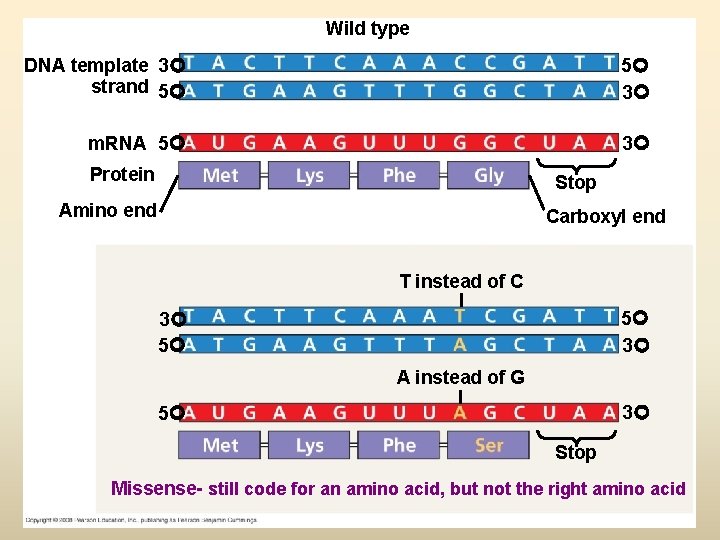
Wild type DNA template 3 strand 5 5 3 m. RNA 5 3 Protein Stop Amino end Carboxyl end T instead of C 5 3 3 5 A instead of G 3 5 Stop Missense- still code for an amino acid, but not the right amino acid
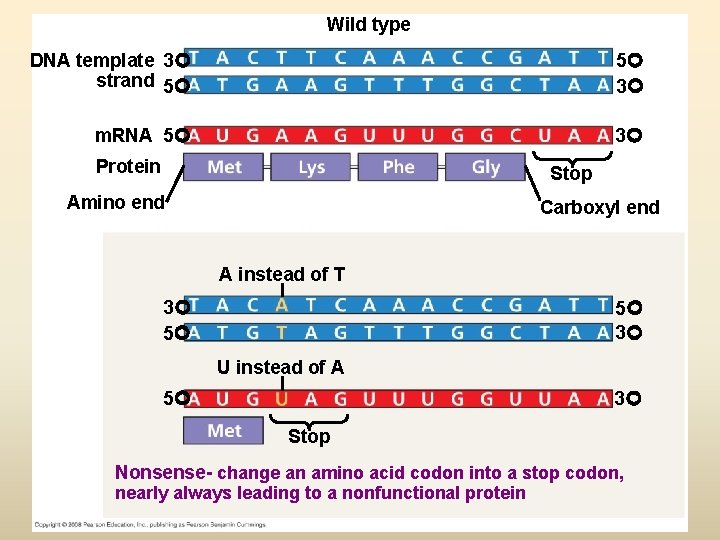
Wild type DNA template 3 strand 5 5 3 m. RNA 5 3 Protein Stop Amino end Carboxyl end A instead of T 3 5 5 3 U instead of A 5 3 Stop Nonsense- change an amino acid codon into a stop codon, nearly always leading to a nonfunctional protein
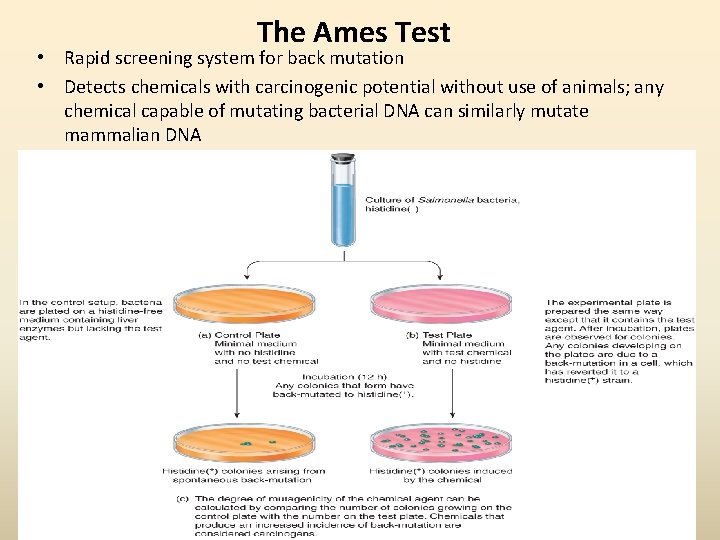
The Ames Test • Rapid screening system for back mutation • Detects chemicals with carcinogenic potential without use of animals; any chemical capable of mutating bacterial DNA can similarly mutate mammalian DNA
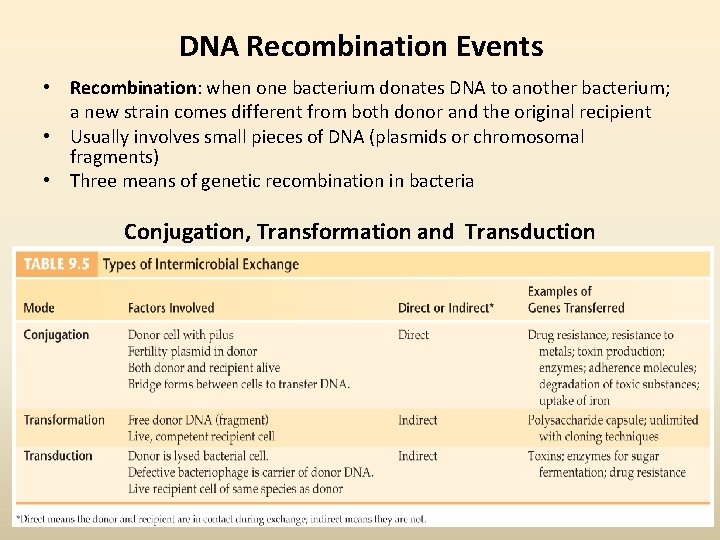
DNA Recombination Events • Recombination: when one bacterium donates DNA to another bacterium; a new strain comes different from both donor and the original recipient • Usually involves small pieces of DNA (plasmids or chromosomal fragments) • Three means of genetic recombination in bacteria Conjugation, Transformation and Transduction
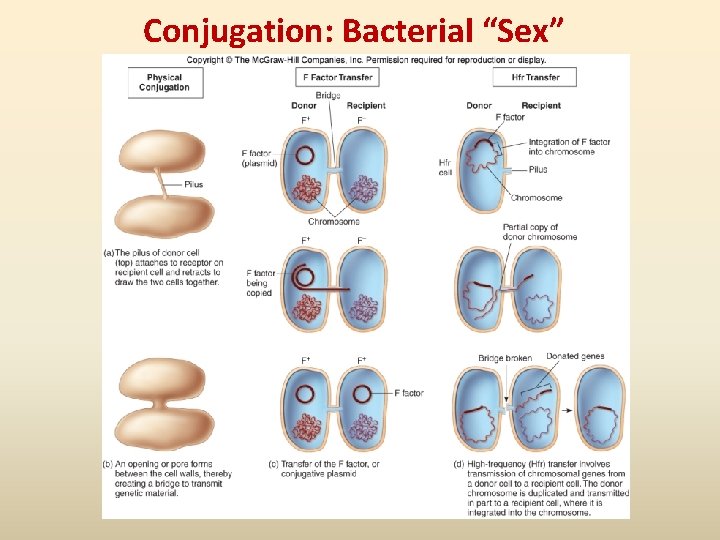
Conjugation: Bacterial “Sex”
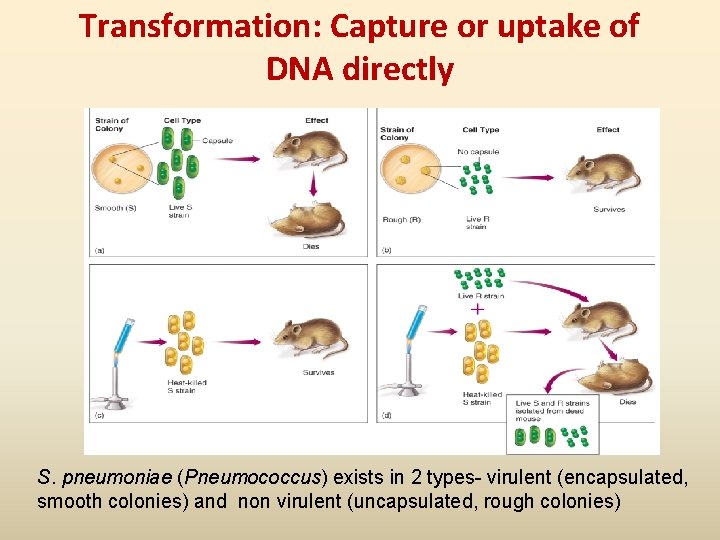
Transformation: Capture or uptake of DNA directly S. pneumoniae (Pneumococcus) exists in 2 types- virulent (encapsulated, smooth colonies) and non virulent (uncapsulated, rough colonies)
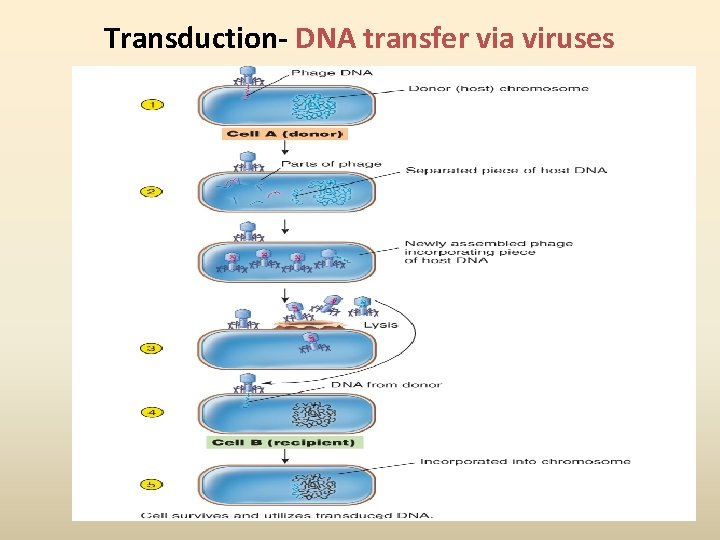
Transduction- DNA transfer via viruses
- Slides: 40