Microbial identification Using API Kits Introduction API Analytical

Microbial identification Using API® Kits
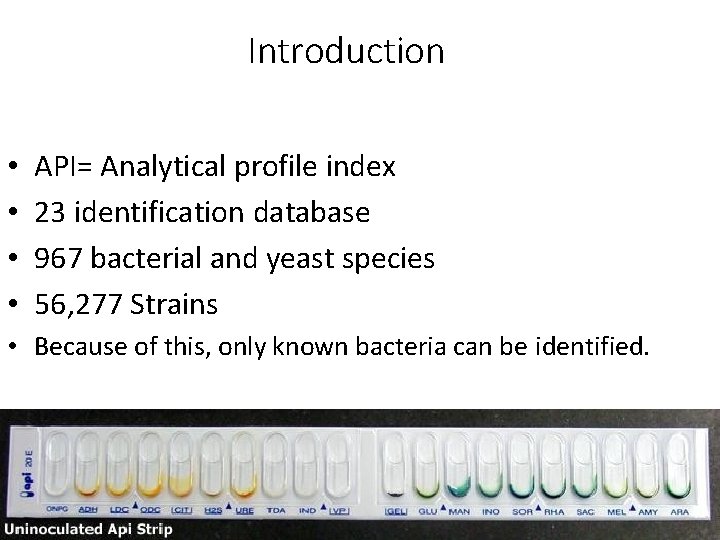
Introduction • • API= Analytical profile index 23 identification database 967 bacterial and yeast species 56, 277 Strains • Because of this, only known bacteria can be identified.
Various identification systems • API® Gram negative Identification • API 20 E – 18 -24 hour identification of Enterobacteriacae and other non-fastidious gram negative bacteria • API Rapid 20 E – 4 -hour identification of Enterobacteriaceae • API 20 NE – 24 to 48 -hour identification of Gram negative non. Enterobacteriaceae • API NH – 2 -hour identification of Neisseria Haemophilus and Branhamella catarrhalis • API Campy – 24 -hour identification of Campylobacter species
Various identification systems • API® Gram positive Identification API Staph – Overnight identification of clinical staphylococci and micrococci • RAPIDEC® Staph – 2 -hour identification of the commonly occurring staphylococci • API 20 Strep – 4 or 24 -hour identification of streptococci and enterococci • API Coryne – 24 -hour identification of Corynebacteria and coryne-like organisms • API Listeria – 24 -hour identification of all Listeria species •
Various identification systems Other API System API® Anaerobe Identification API 20 A®– 24 -hour identification of anaerobes Rapid ID 32 A – 4 -hour identification of anaerobes API® Yeast Identification API 20 C AUX – 48 to 72 -hour identification of yeasts Others • API ® 50 CH – Performance of carbohydrate metabolism tests • API ZYM® – Semi quantitation of enzymatic activities • •
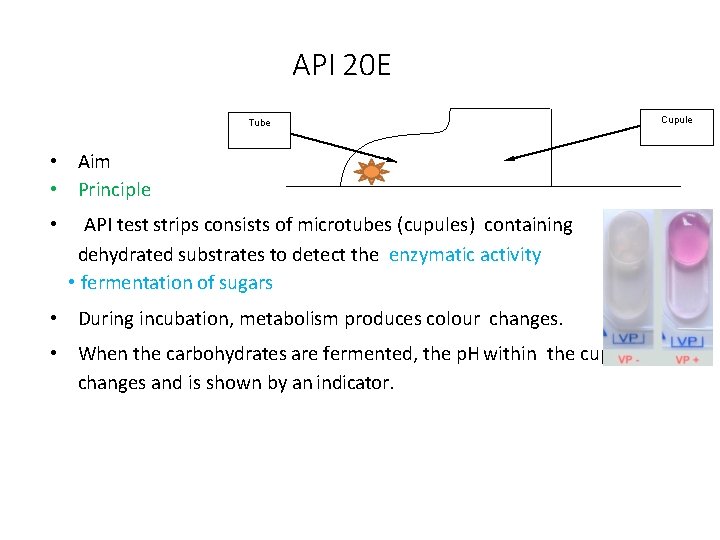
API 20 E Tube • Aim • Principle • API test strips consists of microtubes (cupules) containing dehydrated substrates to detect the enzymatic activity • fermentation of sugars • During incubation, metabolism produces colour changes. • When the carbohydrates are fermented, the p. H within the cupule changes and is shown by an indicator. Cupule
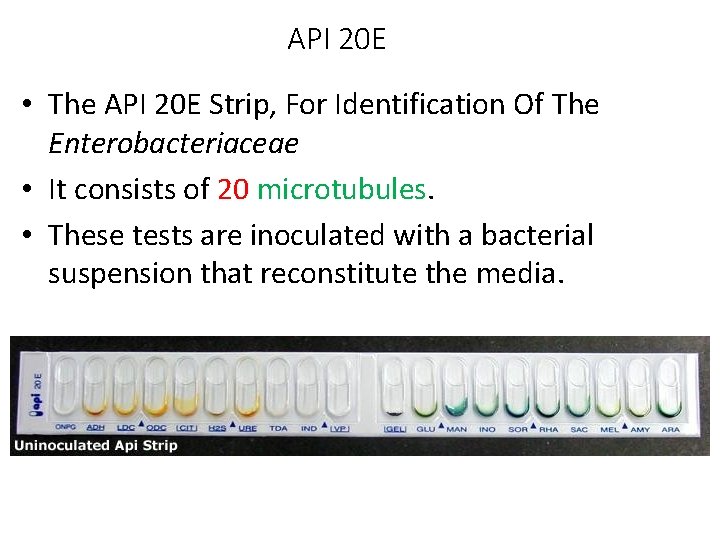
API 20 E • The API 20 E Strip, For Identification Of The Enterobacteriaceae • It consists of 20 microtubules. • These tests are inoculated with a bacterial suspension that reconstitute the media.
API 20 E • • Kit contains API 20 e Strip Media Reagents API strip Trays API code Chart API score Sheet API web/API Catalogue
Analytical Profile Index (API) • Material Needed. 1. API 20 E Strips 2. Incubation boxes 3. Report sheets 4. Disposable Plastic pipettes 5. Disposable plastic inoculating loop 6. 5 ml sterile distilled water 7. Mineral Oil 8. Mac. Conkey agar plate. • Reagent needed. 1. TDA reagent 2. James reagent 3. VPI reagent 4. VP 2 reagent
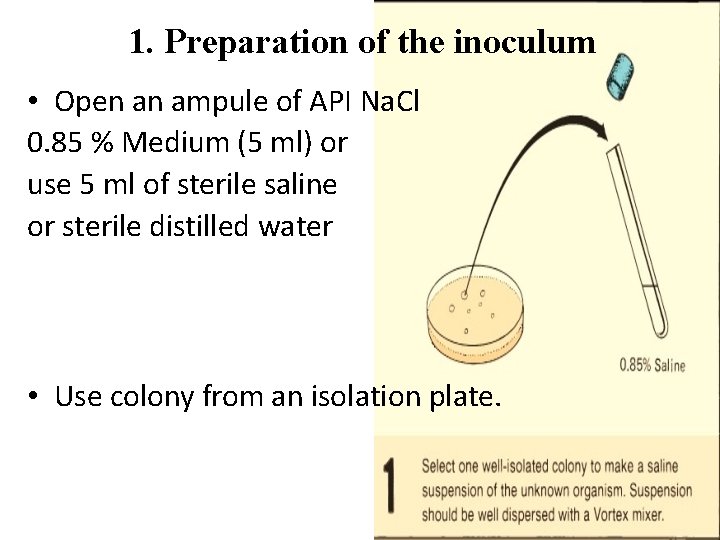
1. Preparation of the inoculum • Open an ampule of API Na. Cl 0. 85 % Medium (5 ml) or use 5 ml of sterile saline or sterile distilled water • Use colony from an isolation plate.
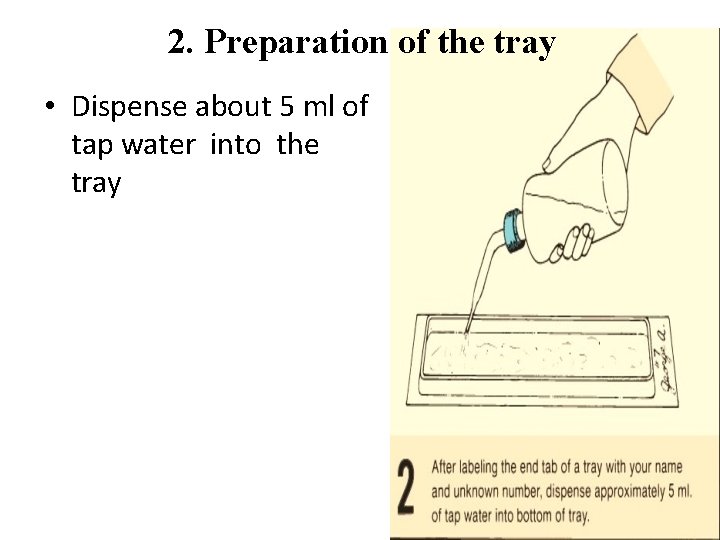
2. Preparation of the tray • Dispense about 5 ml of tap water into the tray
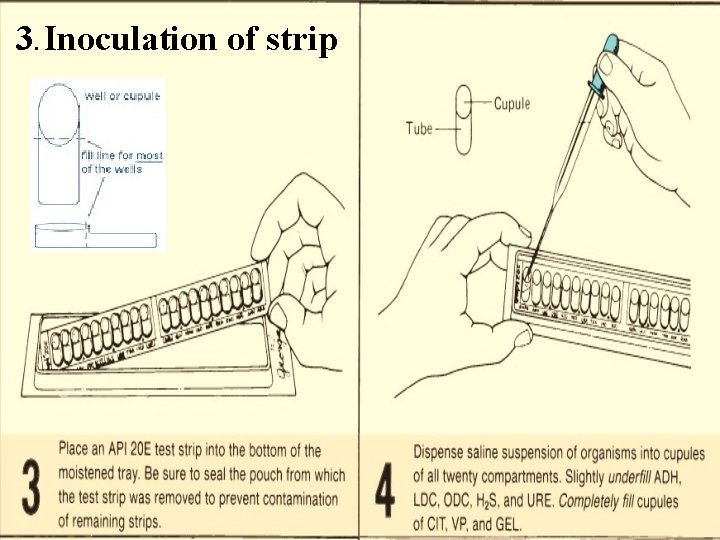
3. Inoculation of strip
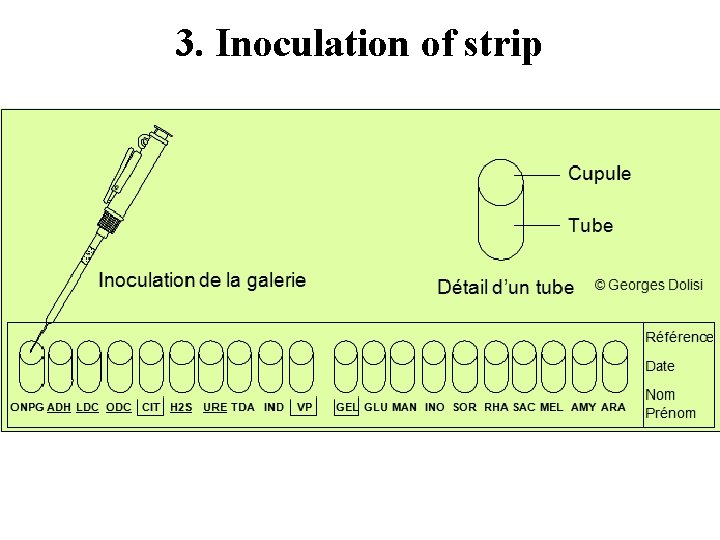
3. Inoculation of strip
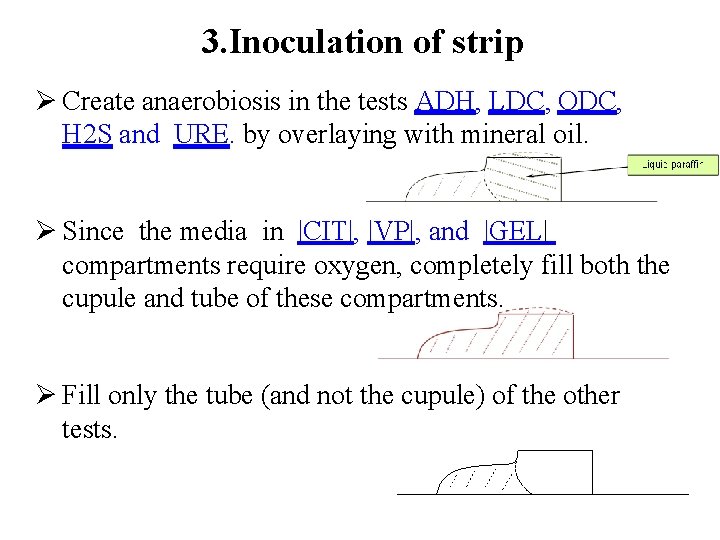
3. Inoculation of strip Create anaerobiosis in the tests ADH, LDC, ODC, H 2 S and URE. by overlaying with mineral oil. Since the media in |CIT|, |VP|, and |GEL| compartments require oxygen, completely fill both the cupule and tube of these compartments. Fill only the tube (and not the cupule) of the other tests.
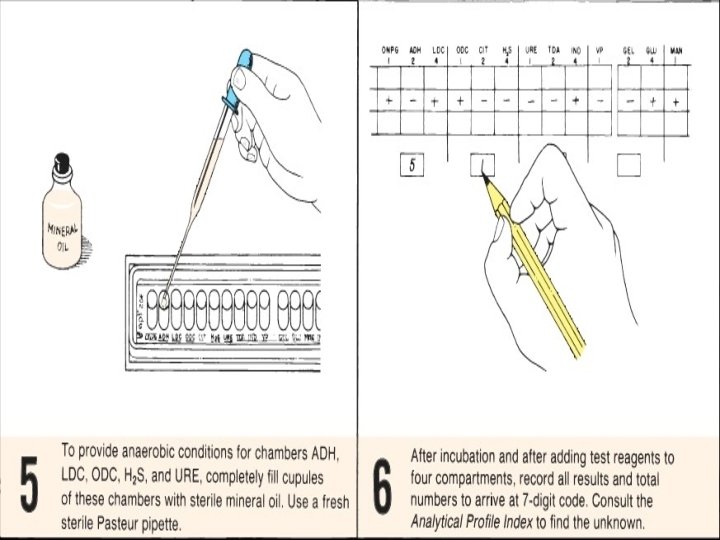

4. Incubation of strip • Inoculate and streak Mac. Conkey purity plate. • Close the incubation box. Incubate the box along with the Mac. Conkey agar at 37°C for 1824 hours.

Results and interpretations • Evaluation of tests, all reactions will be recorded on the laboratory report and test reagents will be added to some compartments. • The seven digit profile number will be determined so the unknown organism can be looked up in the API 20 E analytical profile index. • If the Mac. Conkey purity plate give mix culture, then repeat the test.
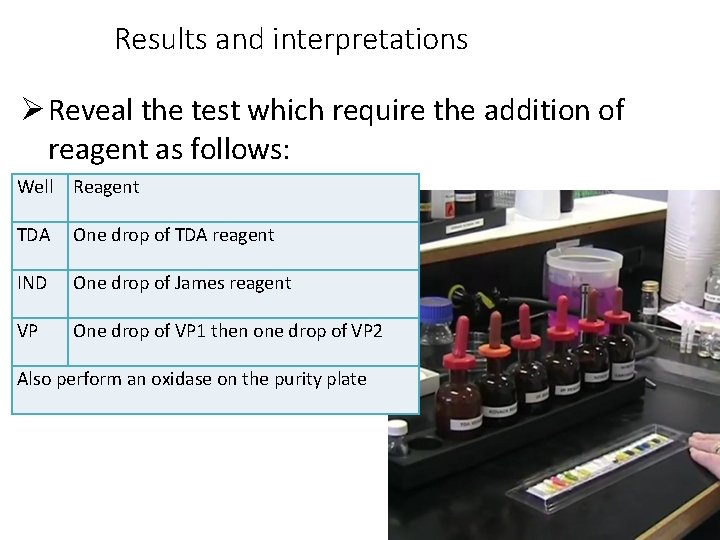
Results and interpretations Reveal the test which require the addition of reagent as follows: Well Reagent TDA One drop of TDA reagent IND One drop of James reagent VP One drop of VP 1 then one drop of VP 2 Also perform an oxidase on the purity plate
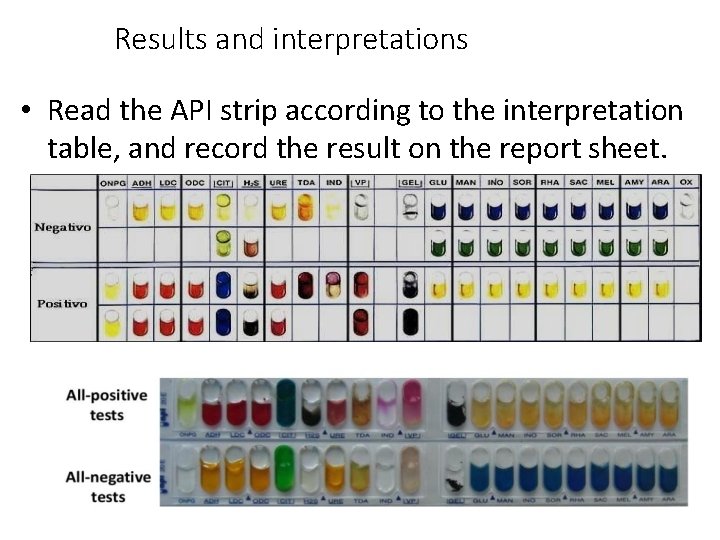
Results and interpretations • Read the API strip according to the interpretation table, and record the result on the report sheet.
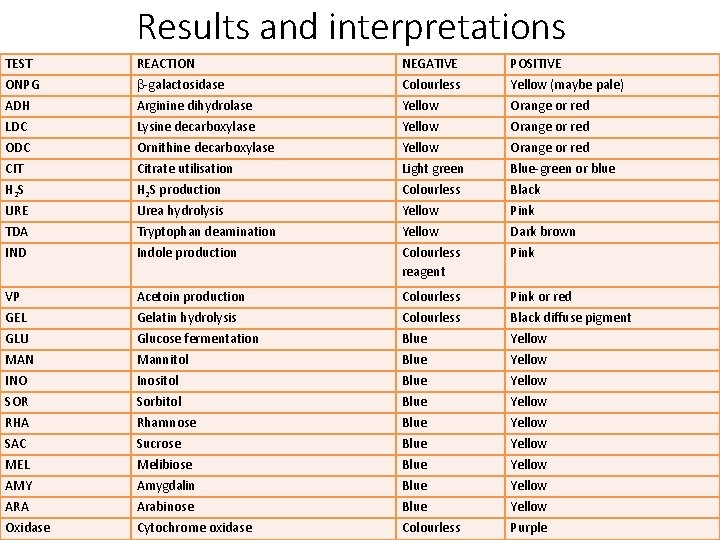
Results and interpretations TEST REACTION NEGATIVE POSITIVE ONPG -galactosidase Colourless Yellow (maybe pale) ADH Arginine dihydrolase Yellow Orange or red LDC Lysine decarboxylase Yellow Orange or red ODC Ornithine decarboxylase Yellow Orange or red CIT Citrate utilisation Light green Blue-green or blue H 2 S production Colourless Black URE Urea hydrolysis Yellow Pink TDA Tryptophan deamination Yellow Dark brown IND Indole production Colourless reagent Pink VP Acetoin production Colourless Pink or red GEL Gelatin hydrolysis Colourless Black diffuse pigment GLU Glucose fermentation Blue Yellow MAN Mannitol Blue Yellow INO Inositol Blue Yellow SOR Sorbitol Blue Yellow RHA Rhamnose Blue Yellow SAC Sucrose Blue Yellow MEL Melibiose Blue Yellow AMY Amygdalin Blue Yellow ARA Arabinose Blue Yellow Oxidase Cytochrome oxidase Colourless Purple

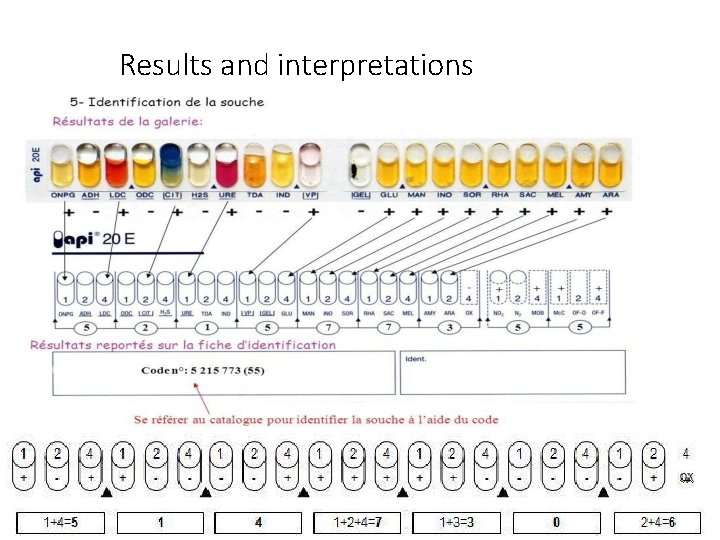
Results and interpretations • On the report sheet, the test are separated into groups of three and number 1 , 2 or 4 is allocated for each test.
Results and interpretations
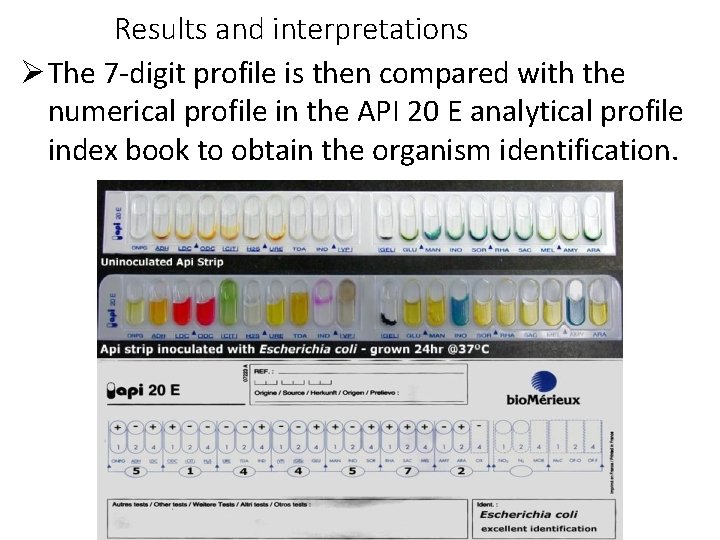
Results and interpretations The 7 -digit profile is then compared with the numerical profile in the API 20 E analytical profile index book to obtain the organism identification.

API Web 1. 3. 0 https: //apiweb. biomerieux. com

Strip Reader and API catalogue
Advantages and Limitations Advantages Easy to use. Economical. Better as compared to traditional biochemical tests. • Rapid Identification • Better identification. • •
Limitations • Based on traditional biochemical testing rather than more sensitive molecular diagnostics. • Results may vary from person to person • Identify only those species that are already present in API database • Interpretation may become difficult at some levels.

Homework • What is the difference between these terms: • Species and strains • sensitivity and specificity

• Question?
- Slides: 29