Microbial Genetics The Structure and Replication of Genomes

Microbial Genetics
The Structure and Replication of Genomes LEARNING OBJECTIVE • Compare and contrast the genome of prokaryotes and eukaryotes • Genetics – Study of inheritance and inheritable traits as expressed in an organism’s genetic material • Genome – The entire genetic complement of an organism – Includes its genes and nucleotide sequences
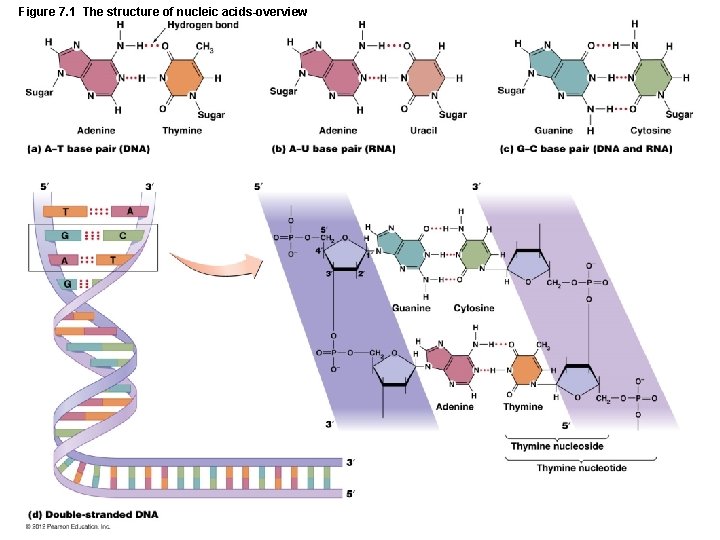
Figure 7. 1 The structure of nucleic acids-overview
The Structure and Replication of Genomes • The Structure of Prokaryotic Genomes – Prokaryotic chromosomes – Main portion of DNA, along with associated proteins and RNA – Prokaryotic cells are haploid (single chromosome copy) – Typical chromosome is circular molecule of DNA in nucleoid
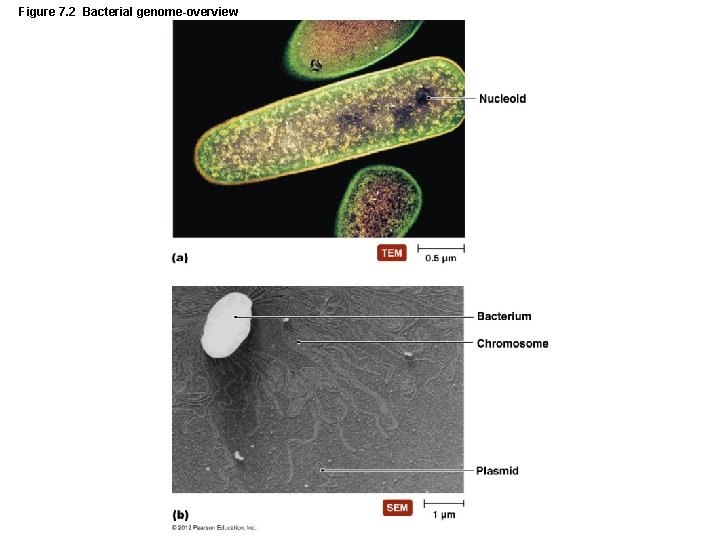
Figure 7. 2 Bacterial genome-overview
The Structure and Replication of Genomes LEARNING OBJECTIVE • Describe the structure and function of Plasmids • The Structure of Prokaryotic Genomes – Plasmids – Small molecules of DNA that replicate independently – Not essential for normal metabolism, growth, or reproduction – Can confer survival advantages – Many types of plasmids – Fertility factors – Resistance factors – Bacteriocin factors – Virulence plasmids
The Structure and Replication of Genomes LEARNING OBJECTIVE • Compare and contrast the genome of prokaryotes and eukaryotes • The Structure of Eukaryotic Genomes – Nuclear chromosomes – Typically have more than one chromosome per cell – Chromosomes are linear and sequestered within nucleus – Eukaryotic cells are often diploid (two chromosome copies)
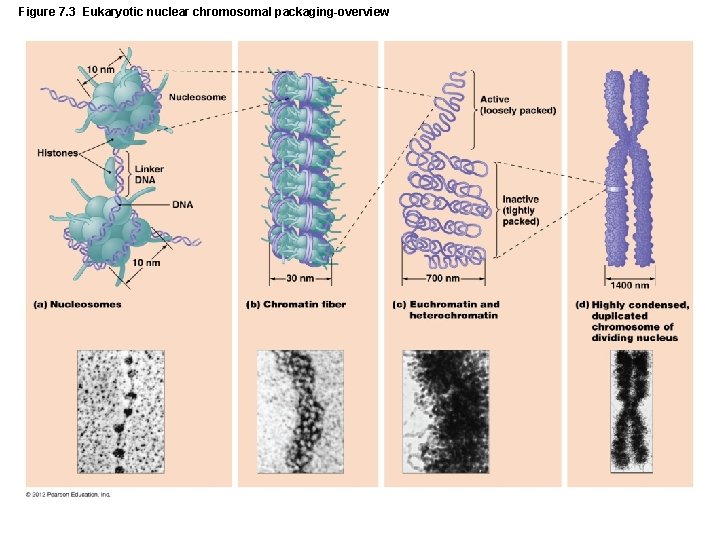
Figure 7. 3 Eukaryotic nuclear chromosomal packaging-overview
The Structure and Replication of Genomes • The Structure of Eukaryotic Genomes – Extranuclear DNA of eukaryotes – DNA molecules of mitochondria and chloroplasts – Resemble chromosomes of prokaryotes – Only code for about 5% of RNA and proteins – Some fungi and protozoa carry plasmids
The Structure and Replication of Genomes LEARNING OBJECTIVE • Describe the replication of DNA as a semicooservative process. • Compare and contrast the synthesis of leading and lagging stands in DNA replication. • DNA Replication – Anabolic polymerization process that requires monomers and energy – Triphosphate deoxyribonucleotides serve both functions – Key to replication is complementary structure of the two strands – Replication is semiconservative – New DNA composed of one original and one daughter strand
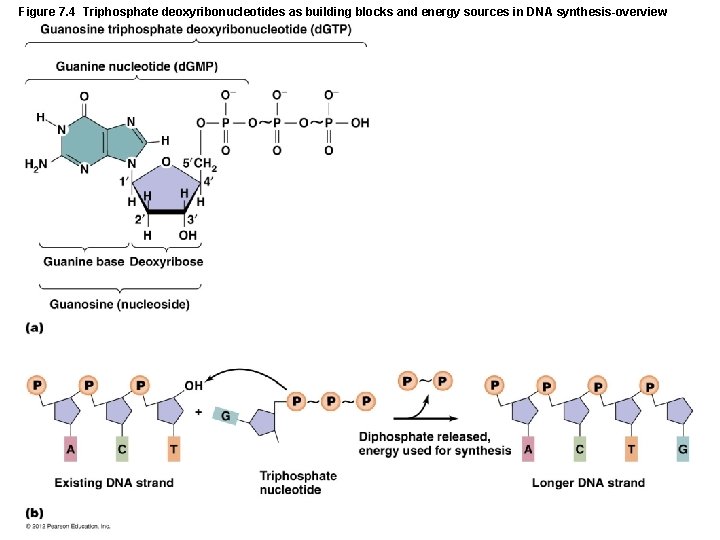
Figure 7. 4 Triphosphate deoxyribonucleotides as building blocks and energy sources in DNA synthesis-overview

Figure 7. 5 Semiconservative model of DNA replication Original DNA First replication Original strand New strands Second replication
The Structure and Replication of Genomes • DNA Replication – Initial processes in replication – Bacterial DNA replication begins at the origin – DNA polymerase replicates DNA only 5 to 3 – Because strands are antiparallel, new strands are synthesized differently – Leading strand synthesized continuously – Lagging strand synthesized discontinuously
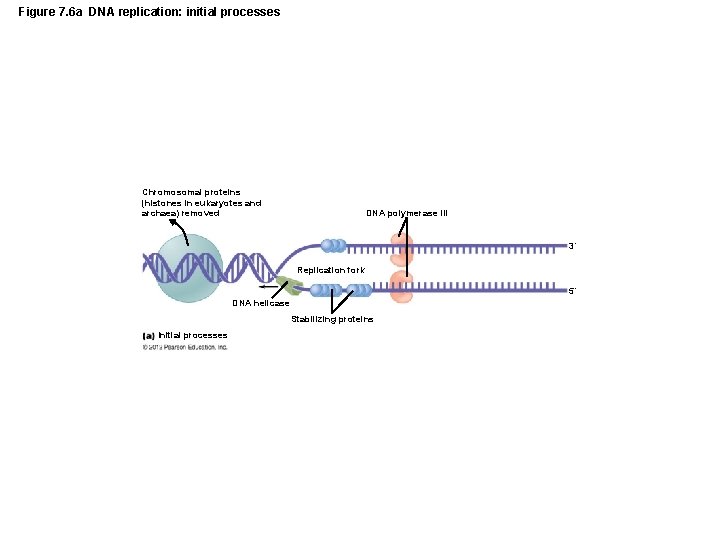
Figure 7. 6 a DNA replication: initial processes Chromosomal proteins (histones in eukaryotes and archaea) removed DNA polymerase III 3´ Replication fork 5´ DNA helicase Stabilizing proteins Initial processes
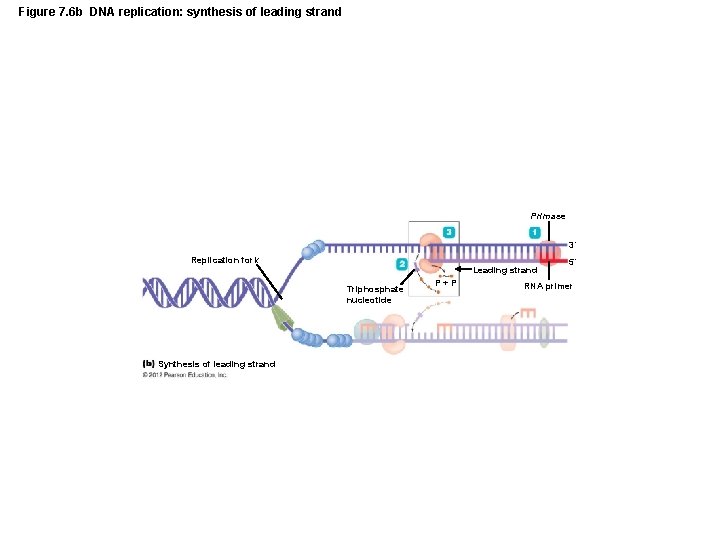
Figure 7. 6 b DNA replication: synthesis of leading strand Primase 3´ Replication fork Leading strand Triphosphate nucleotide Synthesis of leading strand P+P 5´ RNA primer
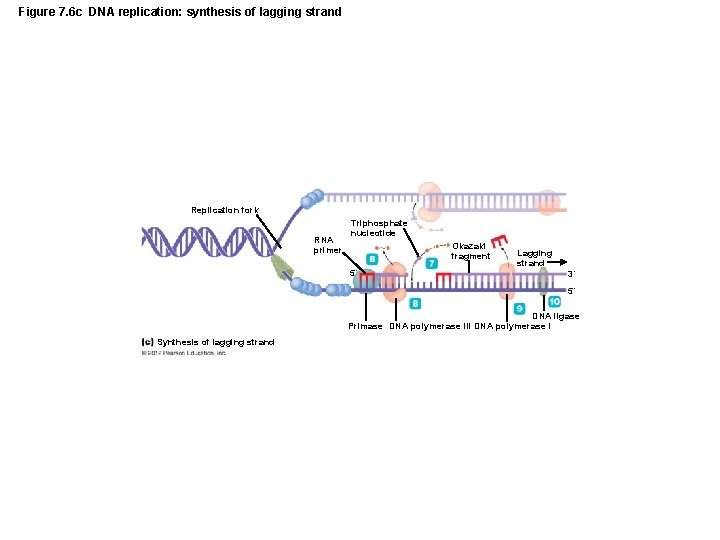
Figure 7. 6 c DNA replication: synthesis of lagging strand Replication fork RNA primer Triphosphate nucleotide Okazaki fragment 5´ Lagging strand 3´ 5´ DNA ligase Primase DNA polymerase III DNA polymerase I Synthesis of lagging strand
The Structure and Replication of Genomes • DNA Replication – Other characteristics of bacterial DNA replication – Bidirectional – Topoisomerases remove supercoils in DNA molecule – DNA is methylated – Control of genetic expression – Initiation of DNA replication – Protection against viral infection – Repair of DNA
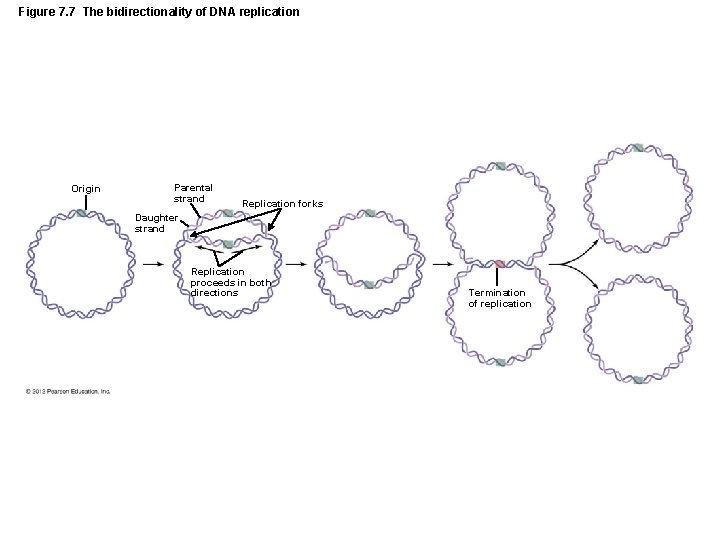
Figure 7. 7 The bidirectionality of DNA replication Origin Parental strand Replication forks Daughter strand Replication proceeds in both directions Termination of replication
The Structure and Replication of Genomes • DNA Replication – Replication of eukaryotic DNA – Similar to bacterial replication – Some differences – Uses four DNA polymerases – Thousands of replication origins – Shorter Okazaki fragments – Plant and animal cells methylate only cytosine bases

Gene Function LEARNING OBJECTIVE • Explain how the genotype of an organism determine its phenotype. • The Relationship Between Genotype and Phenotype – Genotype – Set of genes in the genome – Phenotype – Physical features and functional traits of the organism
Gene Function LEARNING OBJECTIVE • State the central dogma of genetic, and explain the roles of DNA and RNA in polypeptide synthesis. • The Transfer of Genetic Information – Transcription – Information in DNA is copied as RNA – Translation – Polypeptides synthesized from RNA – Central dogma of genetics – DNA transcribed to RNA – RNA translated to form polypeptides
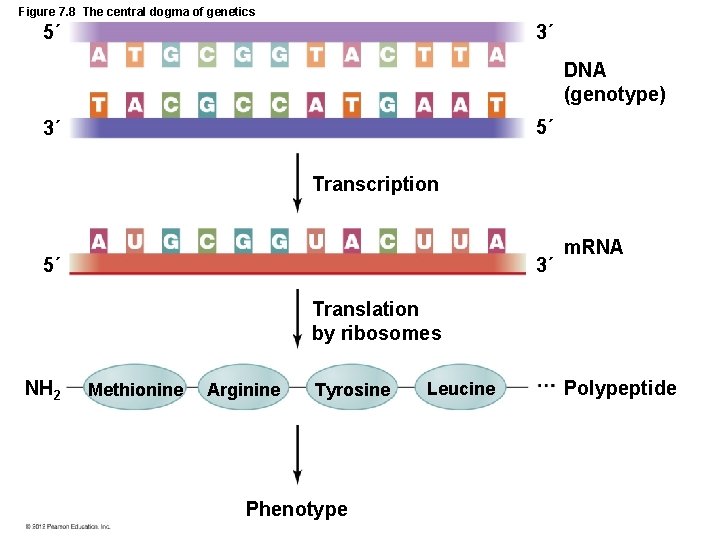
Figure 7. 8 The central dogma of genetics 5´ 3´ DNA (genotype) 5´ 3´ Transcription 5´ 3´ m. RNA Translation by ribosomes NH 2 Methionine Arginine Tyrosine Phenotype Leucine Polypeptide
Gene Function LEARNING OBJECTIVE • Describe three steps in RNA transcription , mentioning the following: DNA , RNA polymerase , promoter, 5’ to 3’ direction and terminator. • The Events in Transcription – Four types of RNA transcribed from DNA – RNA primers – m. RNA – r. RNA – t. RNA – Occur in nucleoid of prokaryotes – Three steps – Initiation – Elongation – Termination
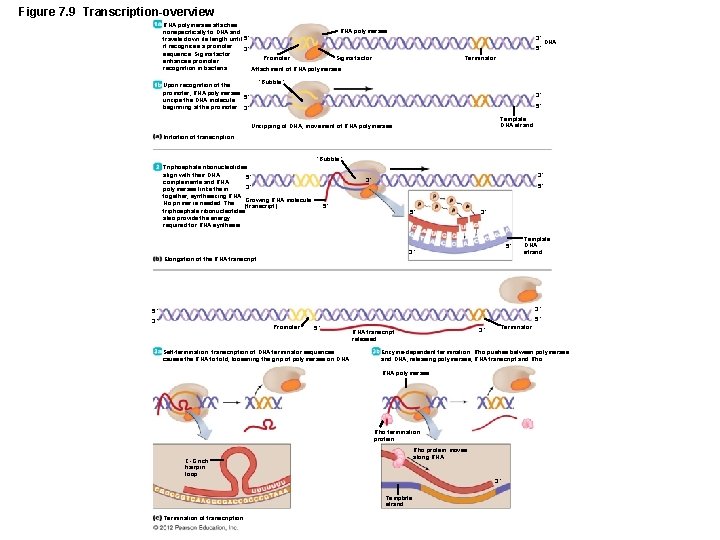
Figure 7. 9 Transcription-overview RNA polymerase attaches RNA polymerase nonspecifically to DNA and travels down its length until 5´ it recognizes a promoter 3´ sequence. Sigma factor Promoter enhances promoter recognition in bacteria. Attachment of RNA polymerase 3´ 5´ DNA Terminator “Bubble” Upon recognition of the promoter, RNA polymerase unzips the DNA molecule 5´ beginning at the promoter. 3´ 3´ 5´ Template DNA strand Unzipping of DNA, movement of RNA polymerase Initiation of transcription “Bubble” Triphosphate ribonucleotides align with their DNA 5´ complements and RNA 3´ polymerase links them together, synthesizing RNA. Growing RNA molecule No primer is needed. The (transcript) triphosphate ribonucleotides also provide the energy required for RNA synthesis. 3´ 3´ 5´ 5´ 5´ 3´ Template DNA strand Elongation of the RNA transcript 3´ 5´ 5´ 3´ Promoter 5´ Self-termination: transcription of DNA terminator sequences causes the RNA to fold, loosening the grip of polymerase on DNA. Terminator 3´ RNA transcript released Enzyme-dependent termination: Rho pushes between polymerase and DNA, releasing polymerase, RNA transcript and Rho. RNA polymerase Rho termination protein Rho protein moves along RNA C-G rich hairpin loop 3´ Template strand Termination of transcription
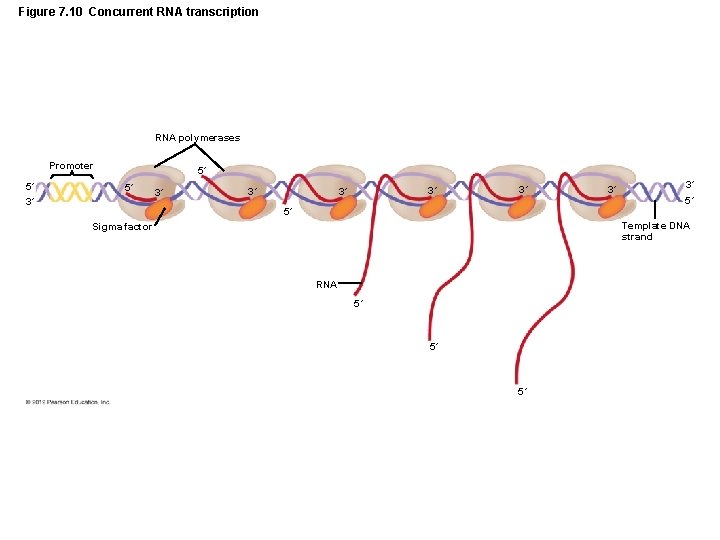
Figure 7. 10 Concurrent RNA transcription RNA polymerases Promoter 5´ 3´ 5´ 5´ 3´ 3´ 3´ 5´ Template DNA strand Sigma factor RNA 5´ 5´ 5´
Gene Function • The Events in Transcription – Transcriptional differences in eukaryotes – RNA transcription occurs in the nucleus – Transcription also occurs in mitochondria and chloroplasts – Three types of RNA polymerases – Numerous transcription factors – m. RNA processed before translation – Capping – Polyadenylation – Splicing
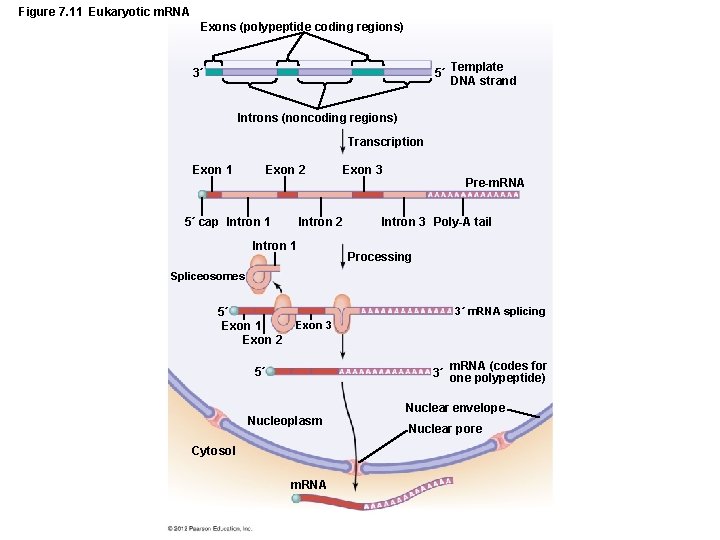
Figure 7. 11 Eukaryotic m. RNA Exons (polypeptide coding regions) 5´ Template DNA strand 3´ Introns (noncoding regions) Transcription Exon 1 Exon 2 5´ cap Intron 1 Intron 2 Intron 1 Exon 3 Pre-m. RNA Intron 3 Poly-A tail Processing Spliceosomes 5´ Exon 1 Exon 2 3´ m. RNA splicing Exon 3 m. RNA (codes for 3´ one polypeptide) 5´ Nucleoplasm Cytosol m. RNA Nuclear envelope Nuclear pore
Gene Function LEARNING OBJECTIVE • Describe the genetic code in general, and identify the relationship between codons and amino acids. • Describe the translation of polypeptides. Identifying the rol of the three type of RNA. • Translation – Process where ribosomes use genetic information of nucleotide sequences to synthesize polypeptides
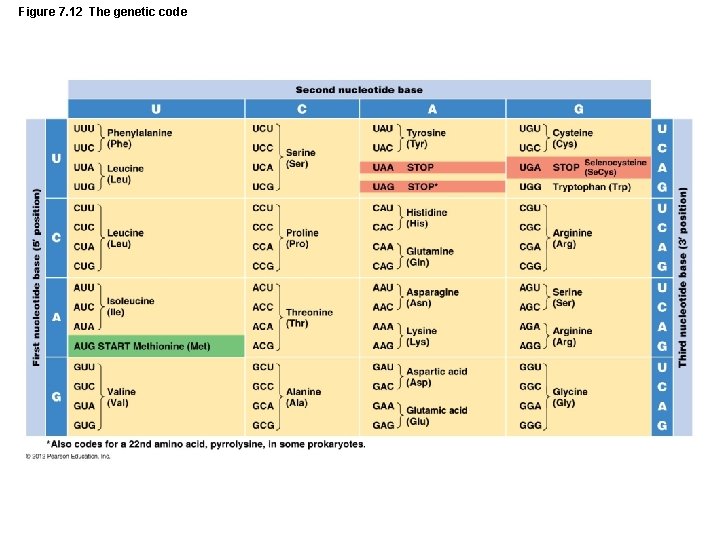
Figure 7. 12 The genetic code
Gene Function • Translation – Participants in translation – Messenger RNA – Transfer RNA – Ribosomes and ribosomal RNA
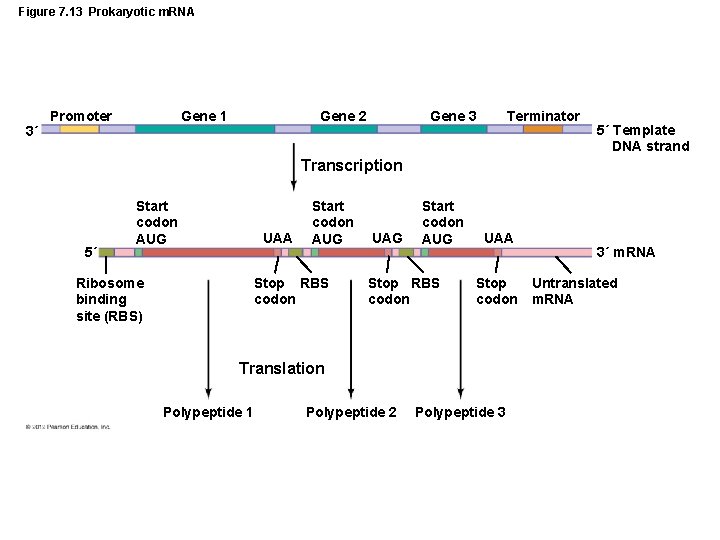
Figure 7. 13 Prokaryotic m. RNA 3´ Promoter Gene 1 Gene 2 Gene 3 Terminator 5´ Template DNA strand Transcription 5´ Start codon AUG UAA Ribosome binding site (RBS) Start codon AUG Stop RBS codon UAG Start codon AUG Stop RBS codon UAA Stop codon Translation Polypeptide 1 Polypeptide 2 Polypeptide 3 3´ m. RNA Untranslated m. RNA
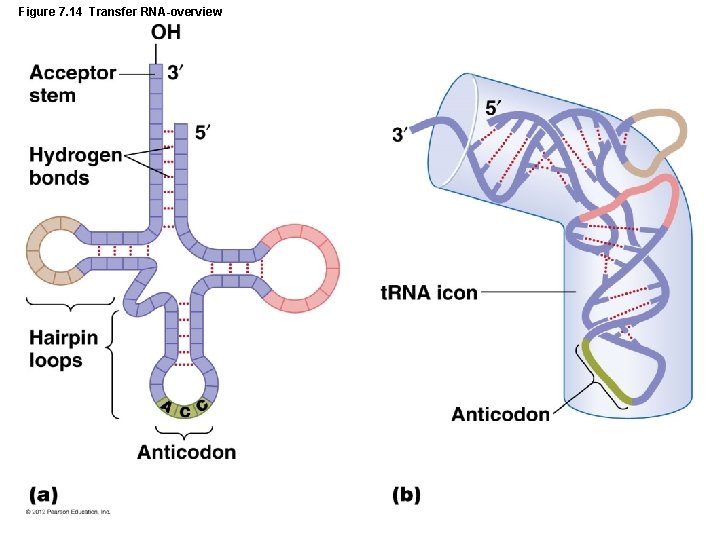
Figure 7. 14 Transfer RNA-overview
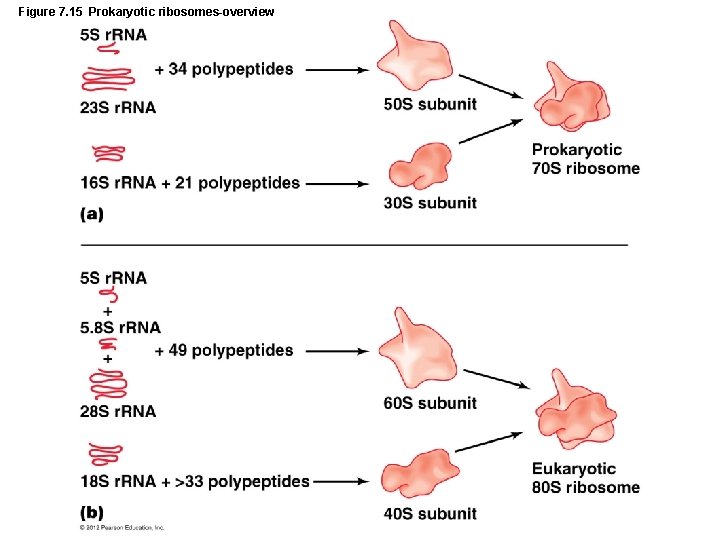
Figure 7. 15 Prokaryotic ribosomes-overview
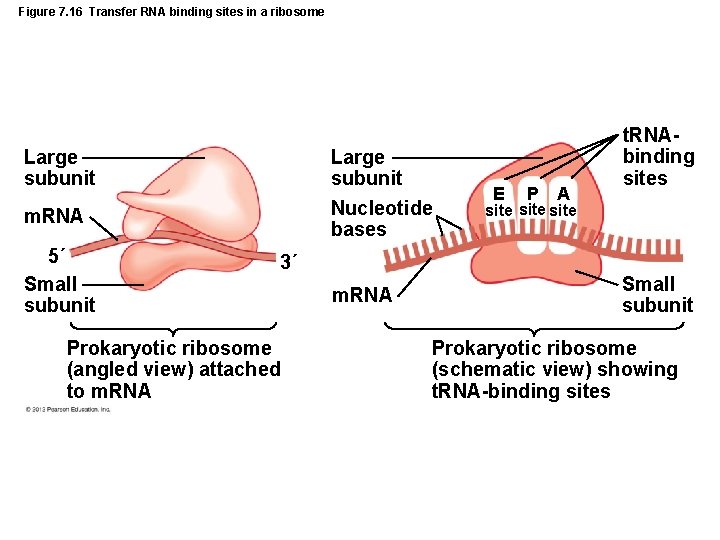
Figure 7. 16 Transfer RNA binding sites in a ribosome Large subunit Nucleotide bases m. RNA 5´ Small subunit 3´ Prokaryotic ribosome (angled view) attached to m. RNA E P A t. RNAbinding sites site Small subunit Prokaryotic ribosome (schematic view) showing t. RNA-binding sites
Gene Function • Translation – Three stages of translation – Initiation – Elongation – Termination – All stages require additional protein factors – Initiation and elongation require energy (GTP)
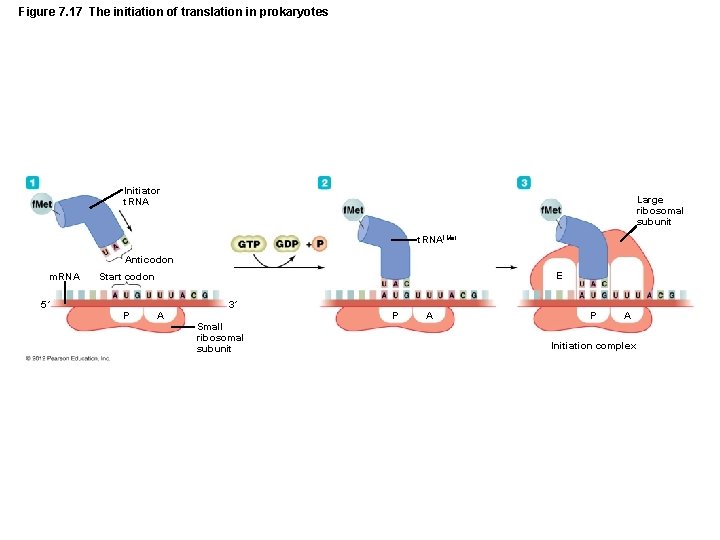
Figure 7. 17 The initiation of translation in prokaryotes Initiator t. RNA Large ribosomal subunit t. RNAf. Met Anticodon m. RNA 5´ E Start codon P A 3´ Small ribosomal subunit P A Initiation complex
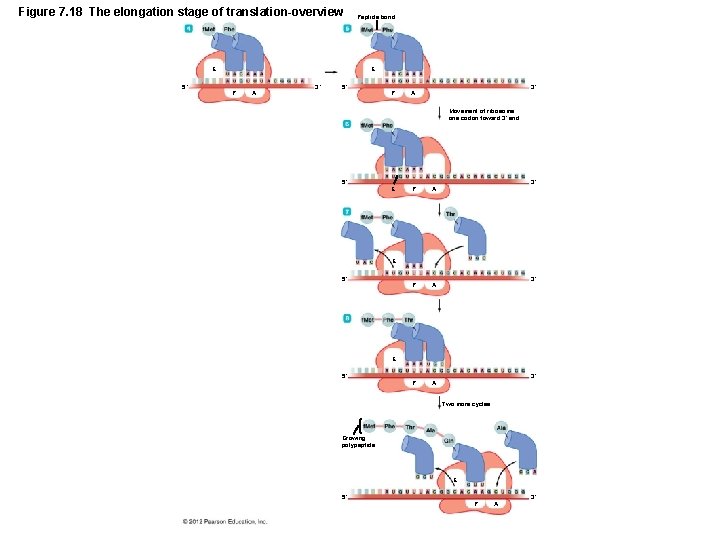
Figure 7. 18 The elongation stage of translation-overview E 5´ Peptide bond E P A 3´ 5´ P 3´ A Movement of ribosome one codon toward 3´ end 5´ E P A P A 3´ E 5´ 3´ Two more cycles Growing polypeptide E 5´ P A 3´
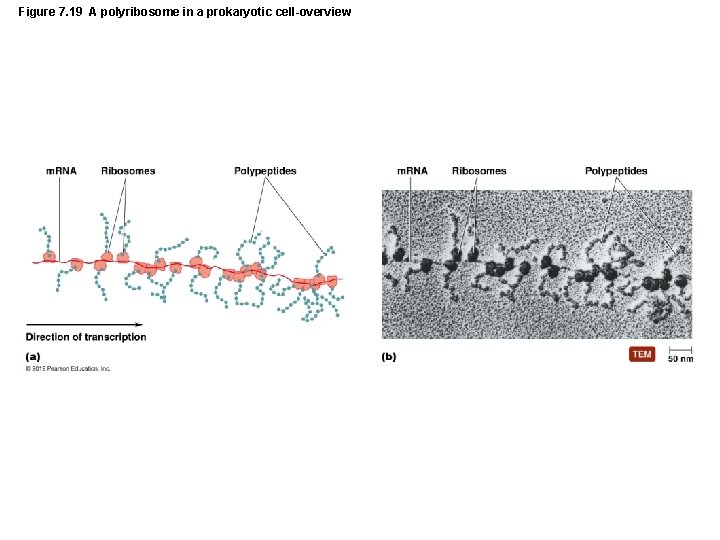
Figure 7. 19 A polyribosome in a prokaryotic cell-overview
Gene Function • Translation – Stages of translation – Termination – Release factors recognize stop codons – Modify ribosome to activate ribozymes – Ribosome dissociates into subunits – Polypeptides released at termination may function alone or together
Gene Function • Translation – Translation differences in eukaryotes – Initiation occurs when ribosomal subunit binds to 5 guanine cap – First amino acid is methionine rather than f-methionine
Gene Function LEARNING OBJECTIVE • Describe the use of ribswitches and short interference RAN in genetic control. • Explain the operon model of transcriptional control in prokaryotes. • Contrast the regulation of an inducible operon with that of a repressible operon , and give an exaple of each. • Regulation of Genetic Expression – 75% of genes are expressed at all times – Other genes transcribed and translated when cells need them – Allows cell to conserve energy – Regulation of protein synthesis – Typically halts transcription – Can stop translation directly
Gene Function • Regulation of Genetic Expression – Control of translation – Genetic expression can be regulated at level of translation – Riboswitch – m. RNA molecule that blocks translation of the polypeptide it encodes – Short interference RNA (si. RNA) – RNA molecule complementary to a portion of m. RNA, t. RNA, or a gene that binds and renders the target inactive
Gene Function • Regulation of Genetic Expression – Nature of prokaryotic operons – An operon consists of a promoter and a series of genes – Some operons are controlled by a regulatory element called an operator
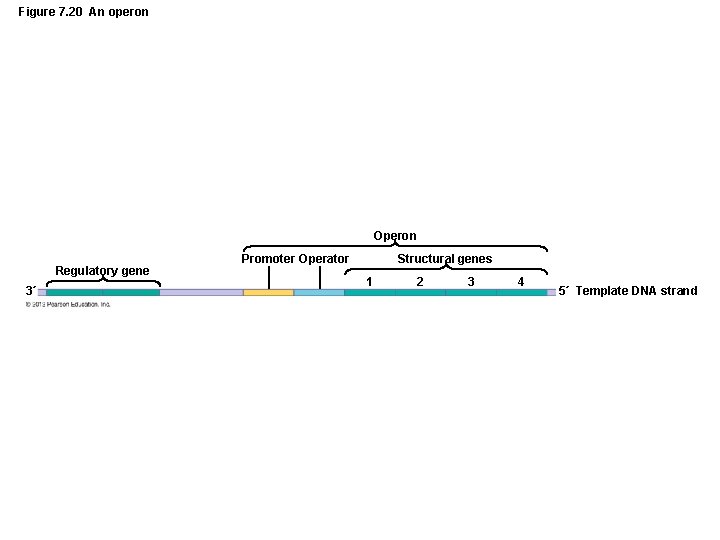
Figure 7. 20 An operon Operon Regulatory gene 3´ Promoter Operator Structural genes 1 2 3 4 5´ Template DNA strand
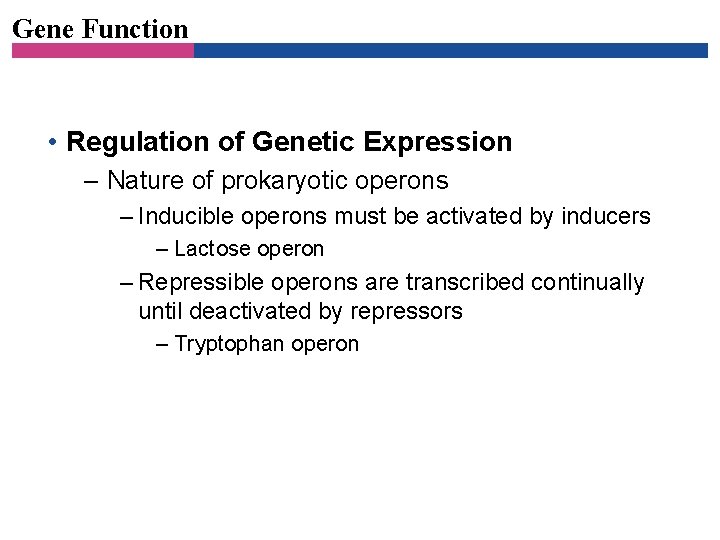
Gene Function • Regulation of Genetic Expression – Nature of prokaryotic operons – Inducible operons must be activated by inducers – Lactose operon – Repressible operons are transcribed continually until deactivated by repressors – Tryptophan operon
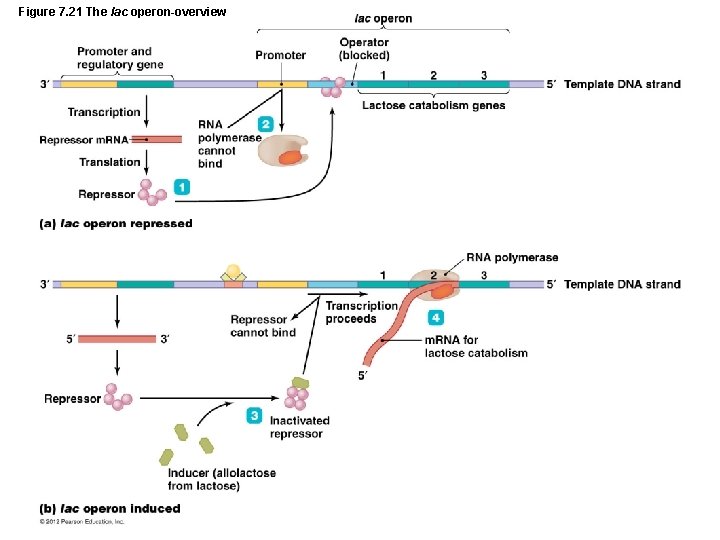
Figure 7. 21 The lac operon-overview
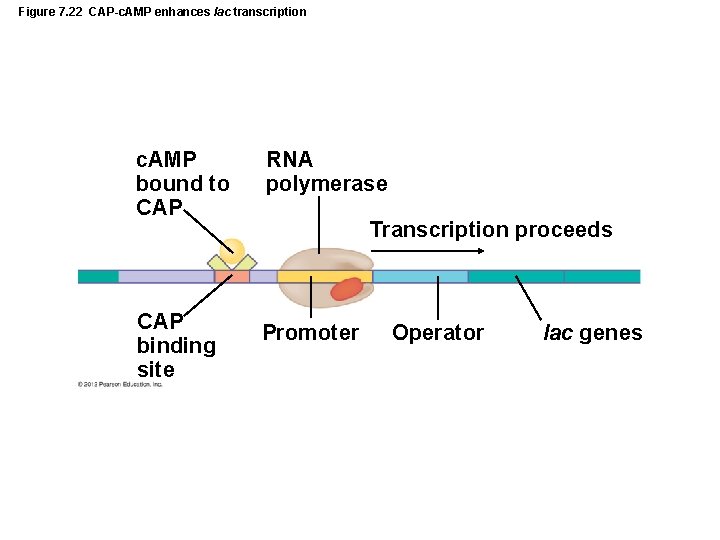
Figure 7. 22 CAP-c. AMP enhances lac transcription c. AMP bound to CAP RNA polymerase CAP binding site Promoter Transcription proceeds Operator lac genes
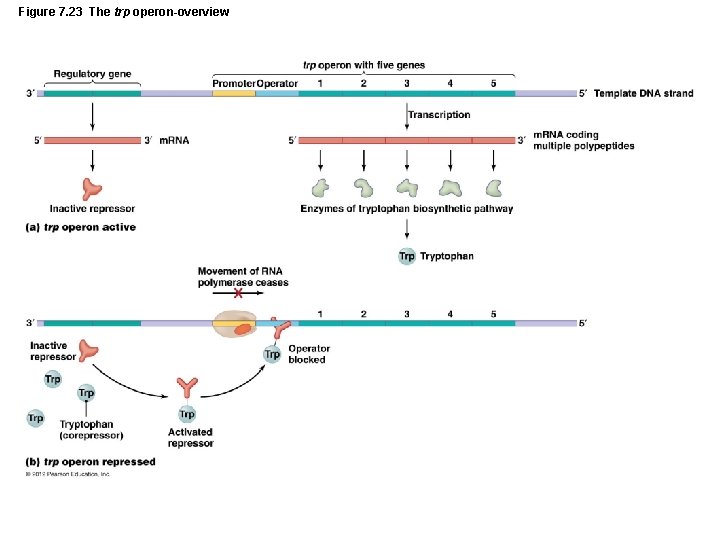
Figure 7. 23 The trp operon-overview
Mutations of Genes LEARNING OBJECTIVE • Define mutation. • Mutation – Change in the nucleotide base sequence of a genome – Rare event – Almost always deleterious – Rarely leads to a protein that improves ability of organism to survive
Mutations of Genes LEARNING OBJECTIVE • Define point mutation and describe three types. • Types of Mutations – Point mutations – Most common – One base pair is affected – Insertions, deletions, and substitutions – Frameshift mutations – Nucleotide triplets after the mutation are displaced – Insertions and deletions

Figure 7. 24 Effects of the various types of point mutations-overview
Mutations of Genes LEARNING OBJECTIVE • Discuss how different types of radiation cause mutation in a genome. • Describe three kind of chemical mutagens and their effects. • Mutagens – Radiation – Ionizing radiation – Nonionizing radiation – Chemical mutagens – Nucleotide analogs – Disrupt DNA and RNA replication – Nucleotide-altering chemicals – Result in base-pair substitutions and missense mutations – Frameshift mutagens – Result in nonsense mutations
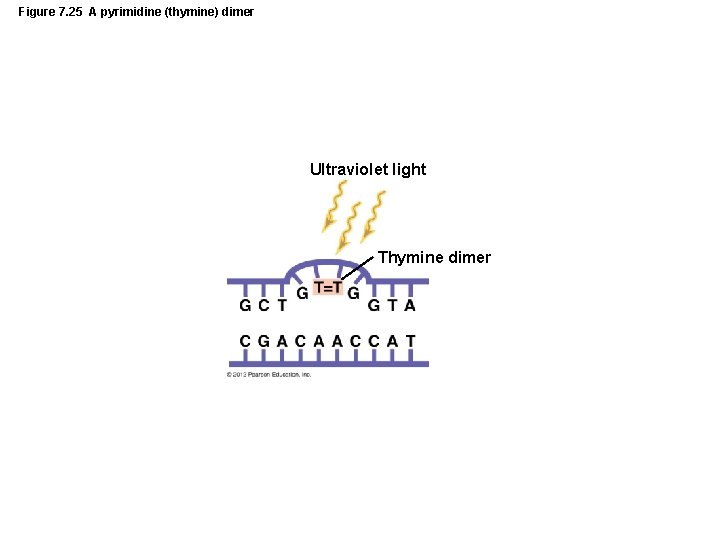
Figure 7. 25 A pyrimidine (thymine) dimer Ultraviolet light Thymine dimer
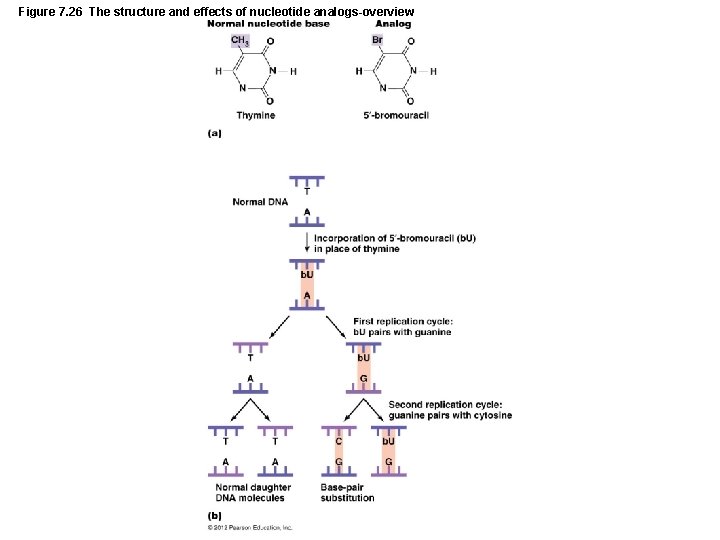
Figure 7. 26 The structure and effects of nucleotide analogs-overview
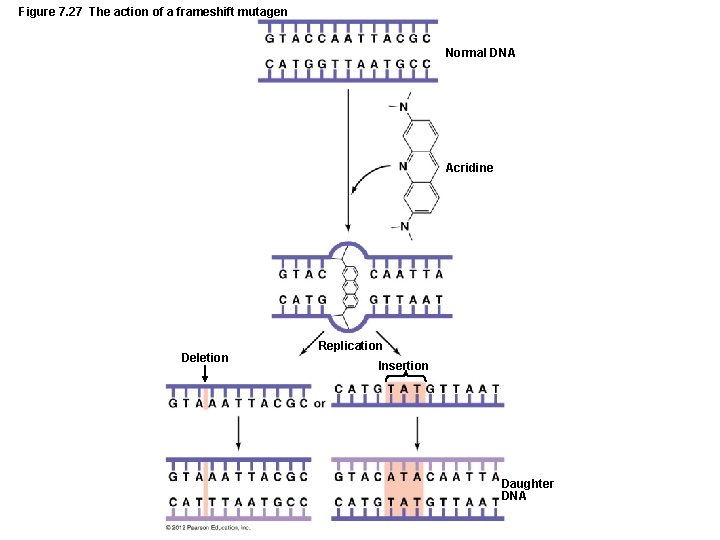
Figure 7. 27 The action of a frameshift mutagen Normal DNA Acridine Deletion Replication Insertion Daughter DNA
Mutations of Genes LEARNING OBJECTIVE • Discuss the relative frequency of deleterious and useful mutations. • Frequency of Mutation – Mutations are rare events – Otherwise organisms could not effectively reproduce – Mutagens increase the mutation rate by a factor of 10 to 1000 times
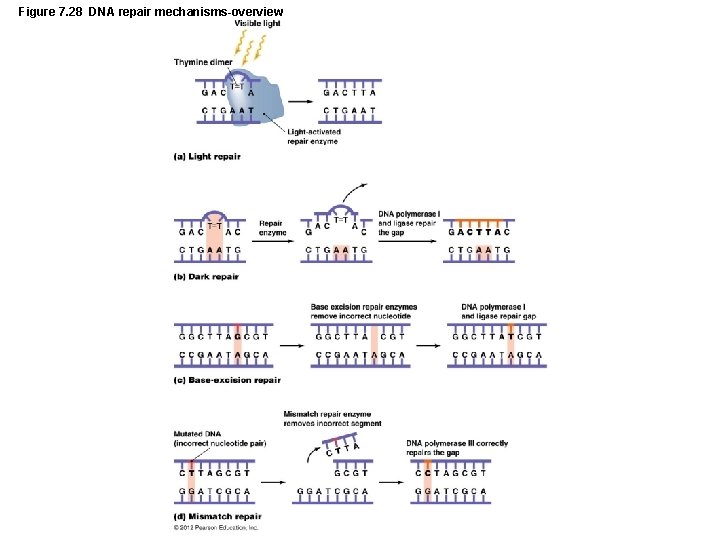
Figure 7. 28 DNA repair mechanisms-overview
Mutations of Genes LEARNING OBJECTIVE • Contrast the positive and negative selection techniques for isolating mutants. • Describe the Ames test , and discuss its use in discovering carcinogens. • Identifying Mutants, Mutagens, and Carcinogens – Mutants – Descendants of a cell that does not repair a mutation – Wild types – Cells normally found in nature – Methods to recognize mutants – Positive selection – Negative (indirect) selection – Ames test
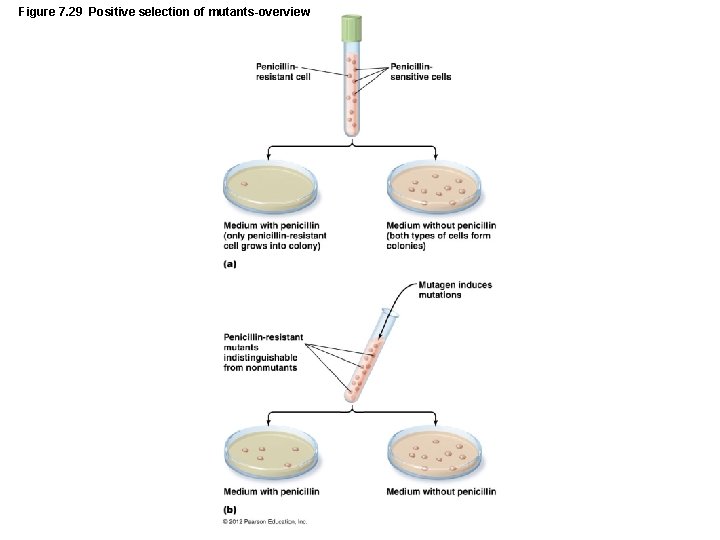
Figure 7. 29 Positive selection of mutants-overview
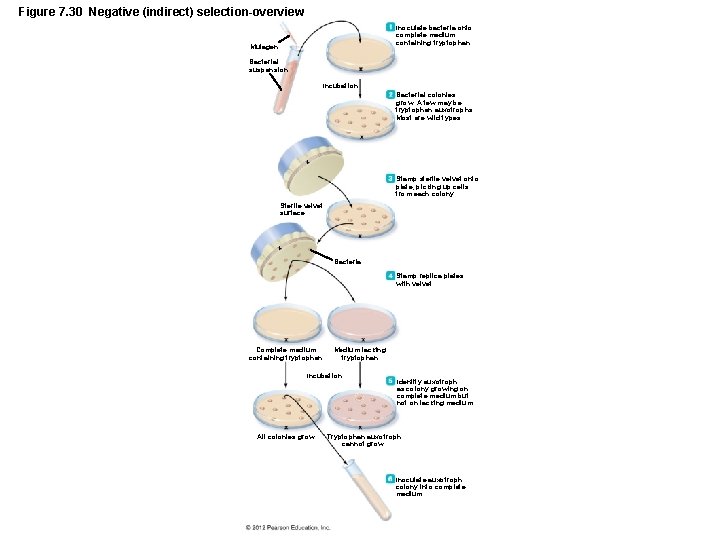
Figure 7. 30 Negative (indirect) selection-overview Inoculate bacteria onto complete medium containing tryptophan. Mutagen Bacterial suspension Incubation Bacterial colonies grow. A few may be tryptophan auxotrophs. Most are wild types. Stamp sterile velvet onto plate, picking up cells from each colony. Sterile velvet surface Bacteria Stamp replica plates with velvet. Complete medium containing tryptophan Medium lacking tryptophan Incubation All colonies grow. Identify auxotroph as colony growing on complete medium but not on lacking medium. Tryptophan auxotroph cannot grow. Inoculate auxotroph colony into complete medium.
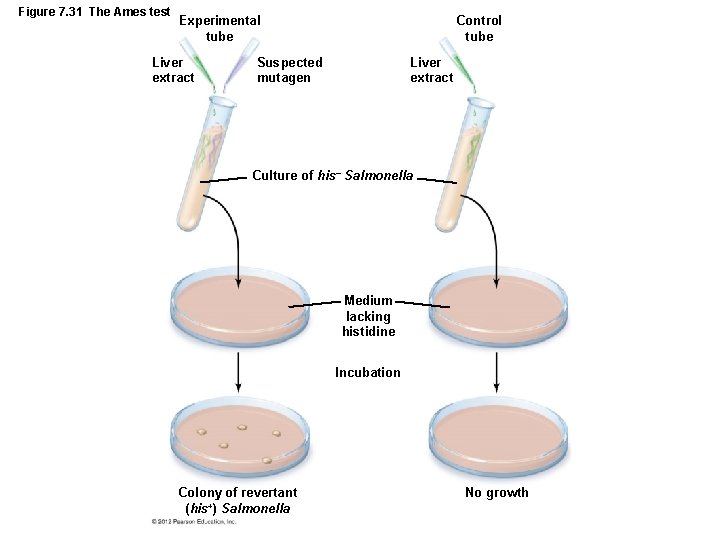
Figure 7. 31 The Ames test Experimental tube Liver extract Control tube Suspected mutagen Liver extract Culture of his– Salmonella Medium lacking histidine Incubation Colony of revertant (his+) Salmonella No growth
Genetic Recombination and Transfer LEARNING OBJECTIVE • Define genetic recombination. • Exchange of nucleotide sequences often mediated by homologous sequences • Recombinants – Cells with DNA molecules that contain new nucleotide sequences • Vertical gene transfer – Organisms replicate their genomes and provide copies to descendants
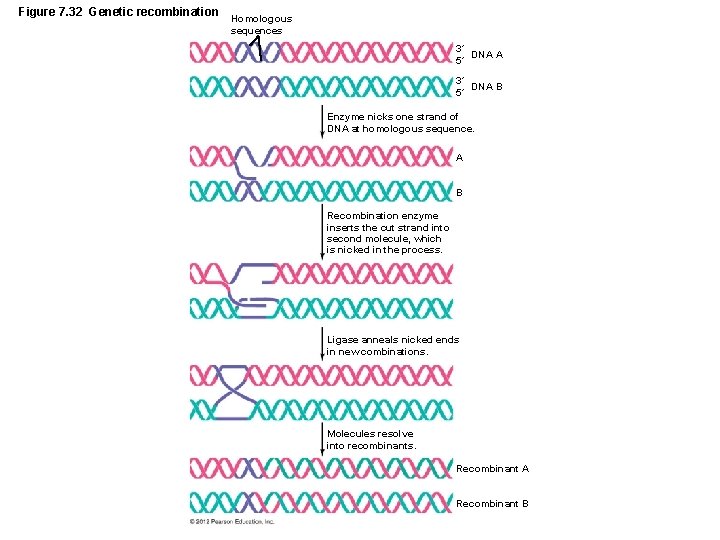
Figure 7. 32 Genetic recombination Homologous sequences 3´ DNA A 5´ 3´ DNA B 5´ Enzyme nicks one strand of DNA at homologous sequence. A B Recombination enzyme inserts the cut strand into second molecule, which is nicked in the process. Ligase anneals nicked ends in new combinations. Molecules resolve into recombinants. Recombinant A Recombinant B
Genetic Recombination and Transfer LEARNING OBJECTIVE • Contrast vertical gene transfer. • Explain the role of an F factor , F+ cell , and Hfr cells in bacterial conjugation. • Describe the structures and action of simple and complex transposons. • Compare and contrast crossing over, transformation, transduction, and conjugation. • Horizontal Gene Transfer Among Prokaryotes – Horizontal gene transfer – Donor cell contributes part of genome to recipient cell – Three types – Transformation – Transduction – Bacterial conjugation
Genetic Recombination and Transfer • Horizontal Gene Transfer Among Prokaryotes – Transformation – One of conclusive pieces of proof that DNA is genetic material – Cells that take up DNA are competent – Results from alterations in cell wall and cytoplasmic membrane that allow DNA to enter cell
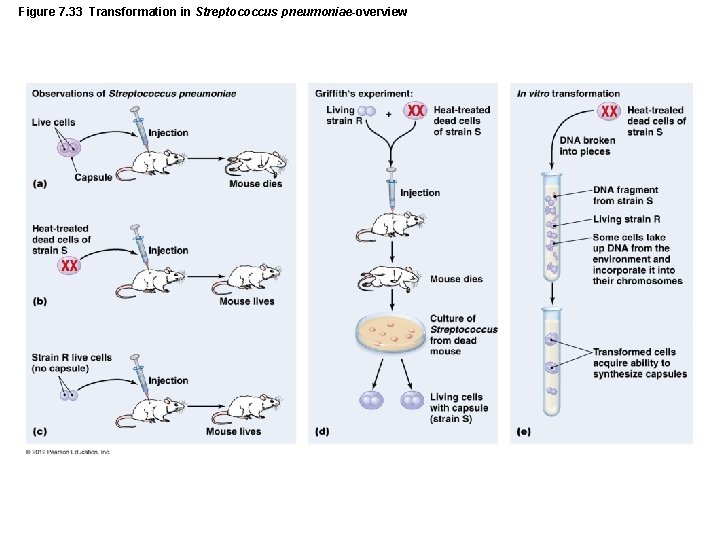
Figure 7. 33 Transformation in Streptococcus pneumoniae-overview
Genetic Recombination and Transfer • Horizontal Gene Transfer Among Prokaryotes – Transduction – Generalized transduction – Transducing phage carries random DNA segment from donor to recipient – Specialized transduction – Only certain donor DNA sequences are transferred
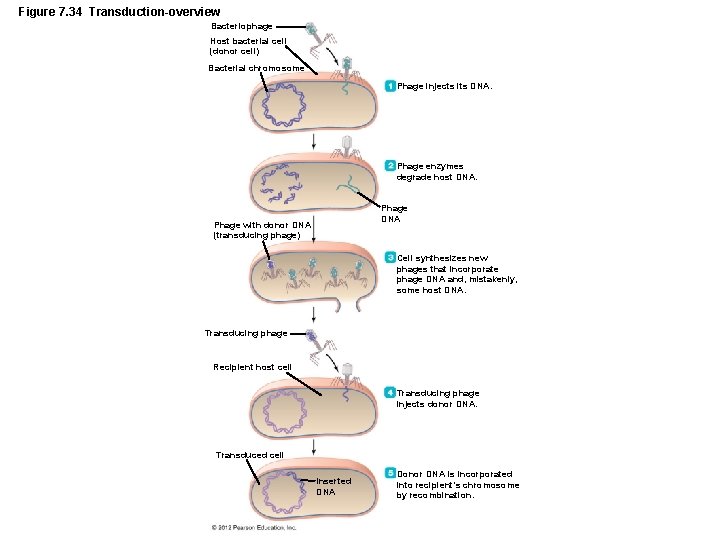
Figure 7. 34 Transduction-overview Bacteriophage Host bacterial cell (donor cell) Bacterial chromosome Phage injects its DNA. Phage enzymes degrade host DNA. Phage DNA Phage with donor DNA (transducing phage) Cell synthesizes new phages that incorporate phage DNA and, mistakenly, some host DNA. Transducing phage Recipient host cell Transducing phage injects donor DNA. Transduced cell Inserted DNA Donor DNA is incorporated into recipient’s chromosome by recombination.
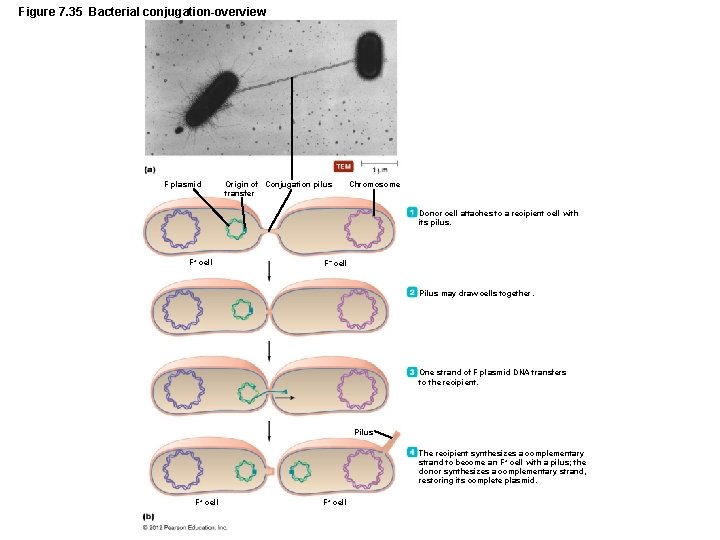
Figure 7. 35 Bacterial conjugation-overview F plasmid Origin of Conjugation pilus transfer Chromosome Donor cell attaches to a recipient cell with its pilus. F+ cell F– cell Pilus may draw cells together. One strand of F plasmid DNA transfers to the recipient. Pilus The recipient synthesizes a complementary strand to become an F + cell with a pilus; the donor synthesizes a complementary strand, restoring its complete plasmid. F+ cell
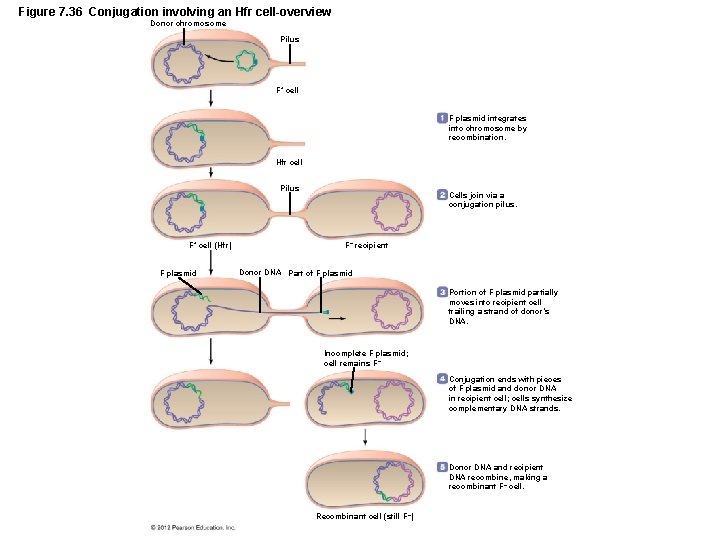
Figure 7. 36 Conjugation involving an Hfr cell-overview Donor chromosome Pilus F+ cell F plasmid integrates into chromosome by recombination. Hfr cell Pilus F+ cell (Hfr) F plasmid Cells join via a conjugation pilus. F– recipient Donor DNA Part of F plasmid Portion of F plasmid partially moves into recipient cell trailing a strand of donor’s DNA. Incomplete F plasmid; cell remains F – Conjugation ends with pieces of F plasmid and donor DNA in recipient cell; cells synthesize complementary DNA strands. Donor DNA and recipient DNA recombine, making a recombinant F– cell. Recombinant cell (still F –)
Genetic Recombination and Transfer • Transposons and Transposition – Transposons – Segments of DNA that move from one location to another in the same or different molecule – Result is a kind of frameshift insertion (transpositions) – Transposons all contain palindromic sequences at each end
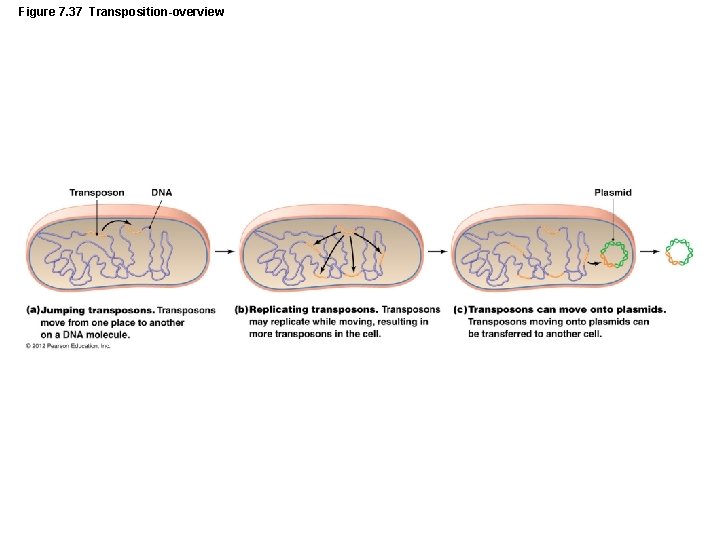
Figure 7. 37 Transposition-overview
Genetic Recombination and Transfer • Transposons and Transposition – Simplest transposons – Insertion sequences – Have no more than two inverted repeats and a gene for transposase – Complex transposons – Contain one or more genes not connected with transposition © 2012 Pearson Education Inc.
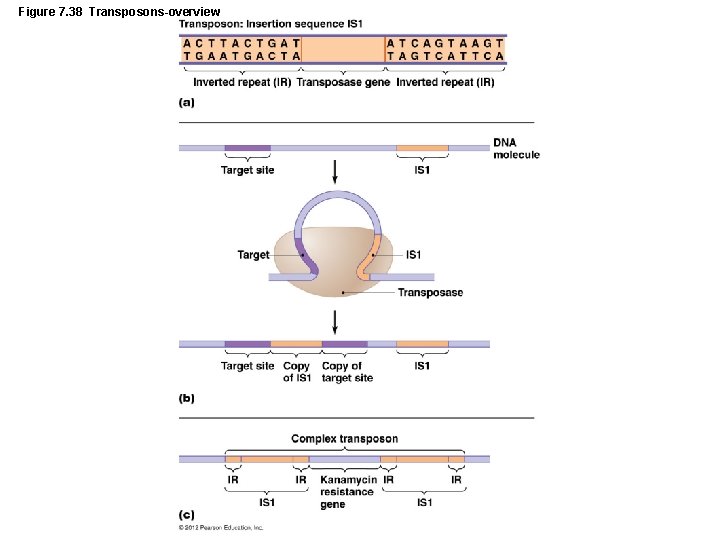
Figure 7. 38 Transposons-overview
- Slides: 74