Microbial Functional Genomics Genomic Technologies And Their Applications

Microbial Functional Genomics, Genomic Technologies, And Their Applications Jizhong (Joe) Zhouj@ornl. gov, 865 -576 -7544 Environmental Sciences Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831, USA 1
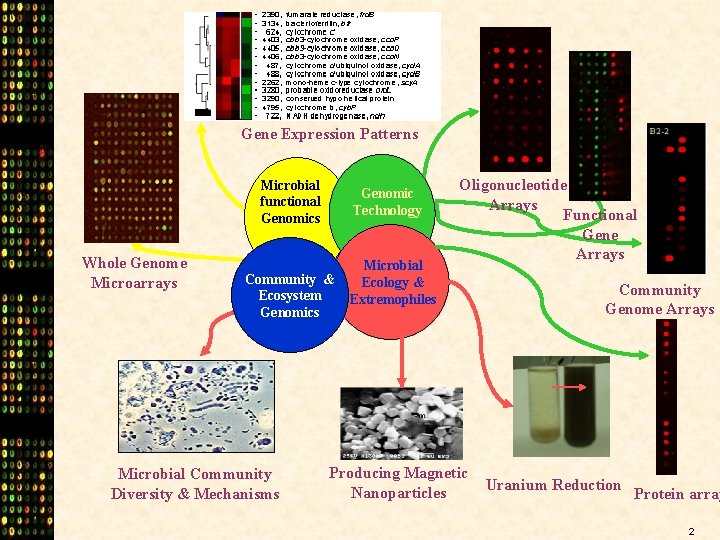
Gene Expression Patterns Microbial functional Genomics Whole Genome Microarrays Genomic Technology Community & Ecosystem Genomics Microbial Community Diversity & Mechanisms Microbial Ecology & Extremophiles Oligonucleotide Arrays Functional Gene Arrays Producing Magnetic Nanoparticles Community Genome Arrays Uranium Reduction Protein array 2
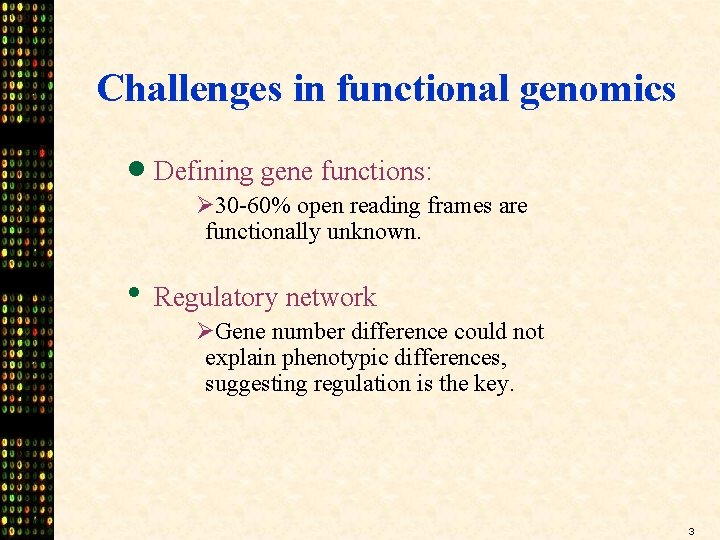
Challenges in functional genomics · Defining gene functions: Ø 30 -60% open reading frames are functionally unknown. • Regulatory network ØGene number difference could not explain phenotypic differences, suggesting regulation is the key. 3
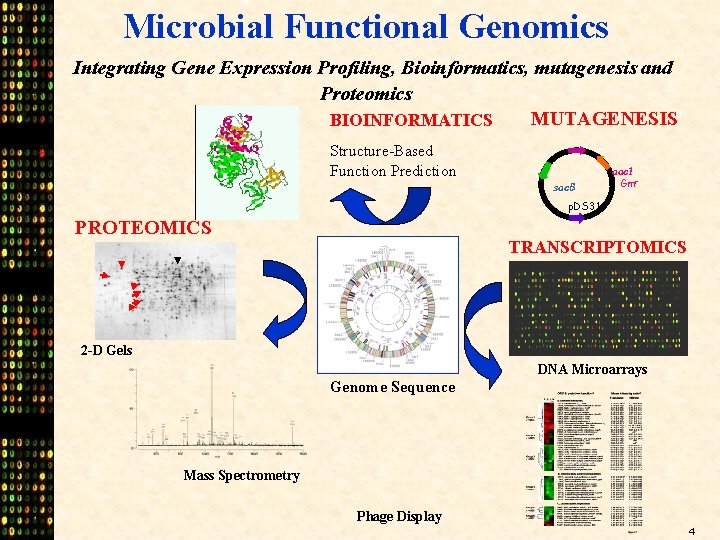
Microbial Functional Genomics Integrating Gene Expression Profiling, Bioinformatics, mutagenesis and Proteomics MUTAGENESIS BIOINFORMATICS Structure-Based Function Prediction sac. B aac 1 Gmr p. DS 31 PROTEOMICS TRANSCRIPTOMICS 2 -D Gels DNA Microarrays Genome Sequence Mass Spectrometry Phage Display 4
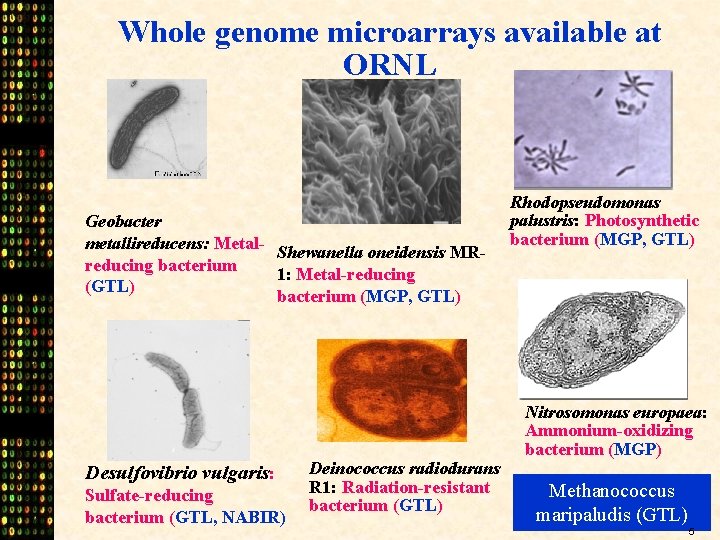
Whole genome microarrays available at ORNL Geobacter metallireducens: Metal. Shewanella oneidensis MRreducing bacterium 1: Metal-reducing (GTL) bacterium (MGP, GTL) Desulfovibrio vulgaris: Sulfate-reducing bacterium (GTL, NABIR) Deinococcus radiodurans R 1: Radiation-resistant bacterium (GTL) Rhodopseudomonas palustris: Photosynthetic bacterium (MGP, GTL) Nitrosomonas europaea: Ammonium-oxidizing bacterium (MGP) Methanococcus maripaludis (GTL) 5
Two primary uses of microarrays for functional analysis • Hypothesis-generating, i. e. , exploratory, Gene expression profiling under different conditions: Øe. g. , Radiation responses in Deinococcus radiodurans. • Hypothesis-driven: Øe. g. , mutant characterization in Shewanella oneidensis MR-1. 6
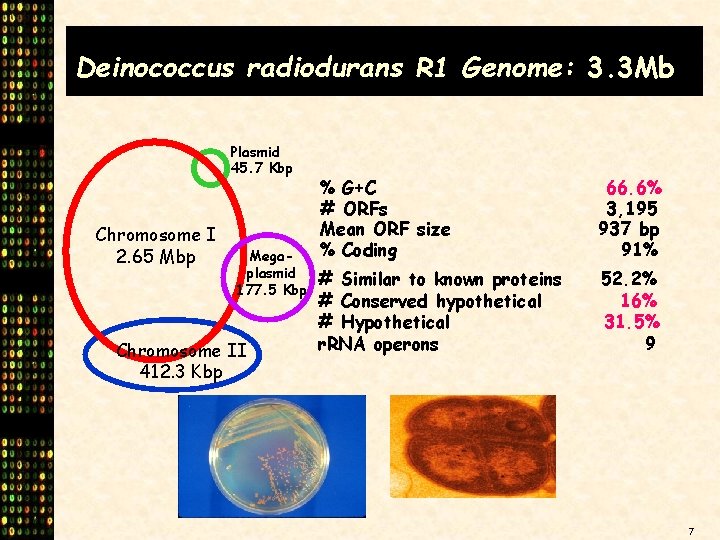
Deinococcus radiodurans R 1 Genome: 3. 3 Mb Plasmid 45. 7 Kbp Chromosome I 2. 65 Mbp Megaplasmid 177. 5 Kbp Chromosome II 412. 3 Kbp % G+C # ORFs Mean ORF size % Coding 66. 6% 3, 195 937 bp 91% # Similar to known proteins # Conserved hypothetical # Hypothetical r. RNA operons 52. 2% 16% 31. 5% 9 *D. radiodurans R 1 genome sequence and annotation courtesy of TIGR 7
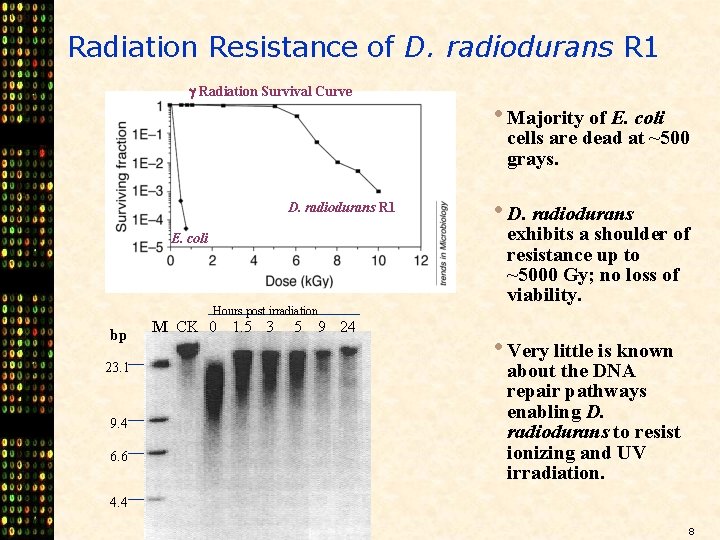
Radiation Resistance of D. radiodurans R 1 Radiation Survival Curve • Majority of E. coli cells are dead at ~500 grays. D. radiodurans R 1 E. coli Hours post irradiation bp 23. 1 9. 4 6. 6 M CK 0 1. 5 3 5 9 24 • D. radiodurans exhibits a shoulder of resistance up to ~5000 Gy; no loss of viability. • Very little is known about the DNA repair pathways enabling D. radiodurans to resist ionizing and UV irradiation. 4. 4 8
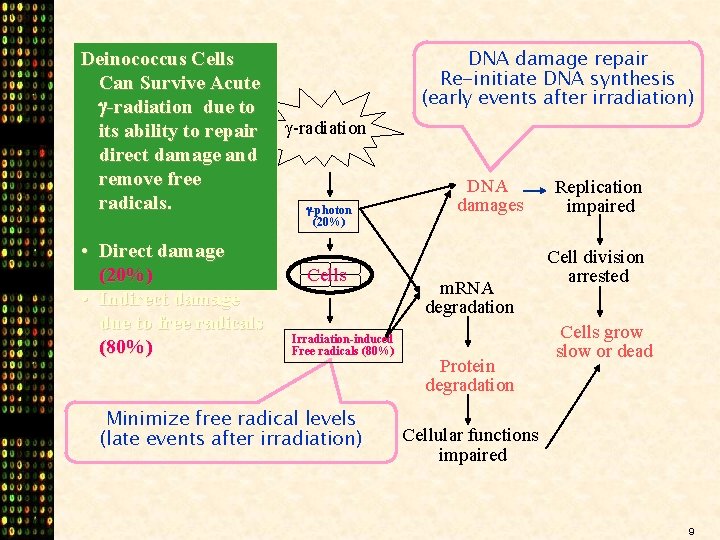
Deinococcus Cells Can Survive Acute -radiation due to its ability to repair direct damage and remove free radicals. • Direct damage (20%) • Indirect damage due to free radicals (80%) DNA damage repair Re-initiate DNA synthesis (early events after irradiation) -radiation -photon (20%) Cells Irradiation-induced Free radicals (80%) Minimize free radical levels (late events after irradiation) DNA damages m. RNA degradation Protein degradation Replication impaired Cell division arrested Cells grow slow or dead Cellular functions impaired 9
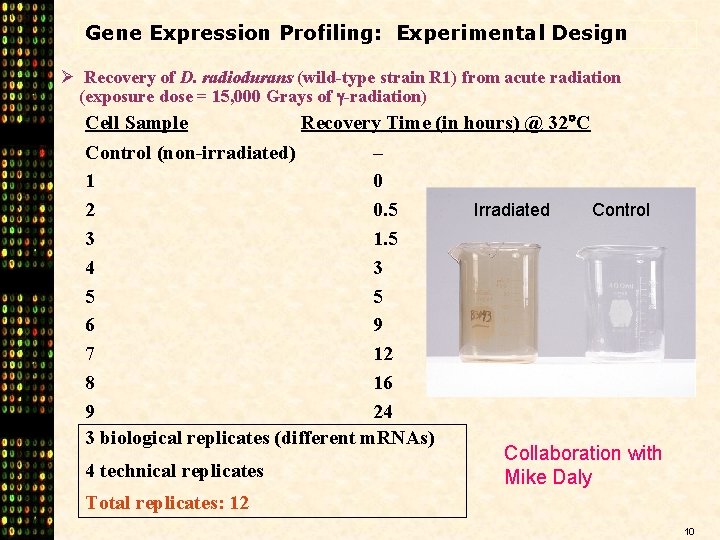
Gene Expression Profiling: Experimental Design Ø Recovery of D. radiodurans (wild-type strain R 1) from acute radiation (exposure dose = 15, 000 Grays of -radiation) Cell Sample Recovery Time (in hours) @ 32 C Control (non-irradiated) – 1 0 Irradiated Control 2 0. 5 3 1. 5 4 3 5 5 6 9 7 12 8 16 9 24 3 biological replicates (different m. RNAs) Collaboration with 4 technical replicates Mike Daly Total replicates: 12 10
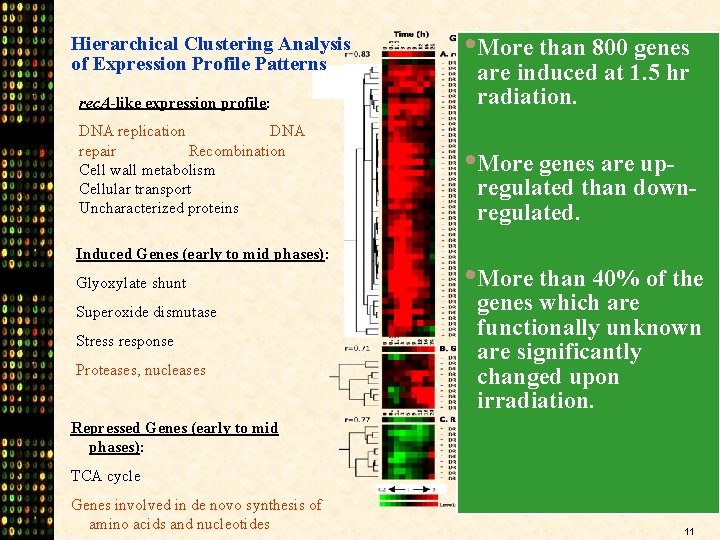
Hierarchical Clustering Analysis of Expression Profile Patterns rec. A-like expression profile: DNA replication DNA repair Recombination Cell wall metabolism Cellular transport Uncharacterized proteins Induced Genes (early to mid phases): Glyoxylate shunt Superoxide dismutase Stress response Proteases, nucleases • More than 800 genes are induced at 1. 5 hr radiation. • More genes are up- regulated than downregulated. • More than 40% of the genes which are functionally unknown are significantly changed upon irradiation. Repressed Genes (early to mid phases): TCA cycle Genes involved in de novo synthesis of amino acids and nucleotides 11
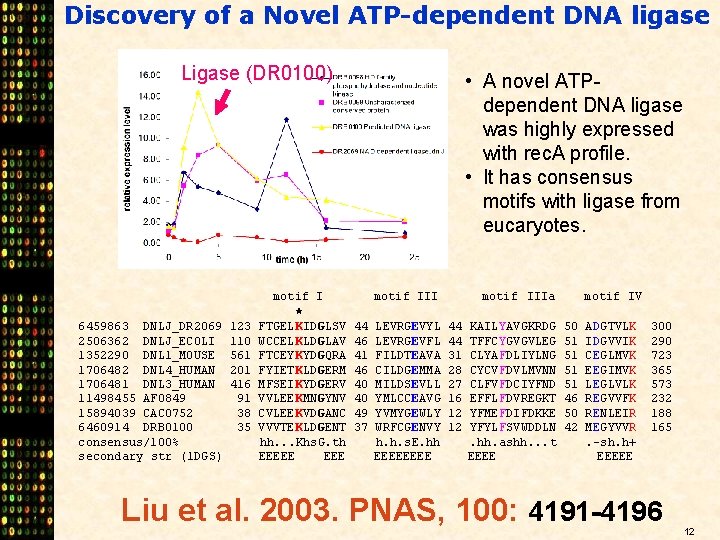
Discovery of a Novel ATP-dependent DNA ligase Ligase (DR 0100) • A novel ATPdependent DNA ligase was highly expressed with rec. A profile. • It has consensus motifs with ligase from eucaryotes. motif IIIa motif IV * 6459863 DNLJ_DR 2069 123 FTGEL KIDGLSV 44 LEVRGEVYL 44 KAILYAVGKRDG 50 ADGTVLK 300 2506362 DNLJ_ECOLI 110 WCCEL KLDGLAV 46 LEVRGEVFL 44 TFFCYGVGVLEG 51 IDGVVIK 290 1352290 DNL 1_MOUSE 561 FTCEY KYDGQRA 41 FILDTEAVA 31 CLYAFDLIYLNG 51 CEGLMVK 723 1706482 DNL 4_HUMAN 201 FYIET KLDGERM 46 CILDGEMMA 28 CYCVFDVLMVNN 51 EEGIMVK 365 1706481 DNL 3_HUMAN 416 MFSEI KYDGERV 40 MILDSEVLL 27 CLFVFDCIYFND 51 LEGLVLK 573 11498455 AF 0849 91 VVLEE KMNGYNV 40 YMLCCEAVG 16 EFFLFDVREGKT 46 REGVVFK 232 15894039 CAC 0752 38 CVLEE KVDGANC 49 YVMYGEWLY 12 YFMEFDIFDKKE 50 RENLEIR 188 6460914 DRB 0100 35 VVVTE KLDGENT 37 WRFCGENVY 12 YFYLFSVWDDLN 42 MEGYVVR 165 consensus/100% hh. . . Khs. G. th h. h. s. E. hh . hh. ashh. . . t . -sh. h+ secondary str (1 DGS) EEEEE EEEE EEEEE Liu et al. 2003. PNAS, 100: 4191 -4196 12
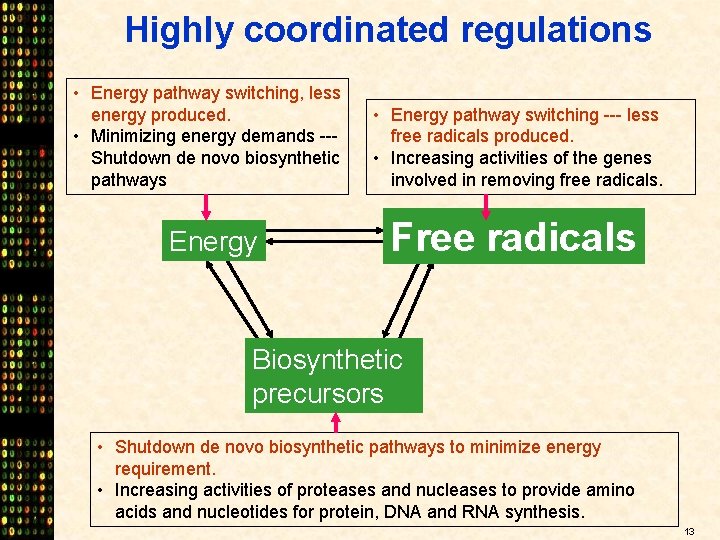
Highly coordinated regulations • Energy pathway switching, less energy produced. • Minimizing energy demands --Shutdown de novo biosynthetic pathways Energy • Energy pathway switching --- less free radicals produced. • Increasing activities of the genes involved in removing free radicals. Free radicals Biosynthetic precursors • Shutdown de novo biosynthetic pathways to minimize energy requirement. • Increasing activities of proteases and nucleases to provide amino acids and nucleotides for protein, DNA and RNA synthesis. 13
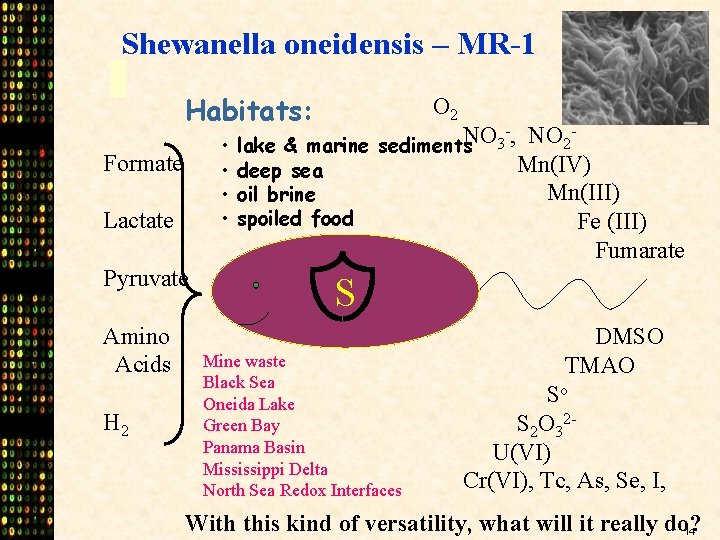
Shewanella oneidensis – MR-1 Habitats: • • Formate Lactate Pyruvate Amino Acids H 2 O 2 -, NO NO 2 lake & marine sediments 3 Mn(IV) deep sea Mn(III) oil brine spoiled food Fe (III) Fumarate S Mine waste Black Sea Oneida Lake Green Bay Panama Basin Mississippi Delta North Sea Redox Interfaces DMSO TMAO So S 2 O 32 U(VI) Cr(VI), Tc, As, Se, I, With this kind of versatility, what will it really do? 14
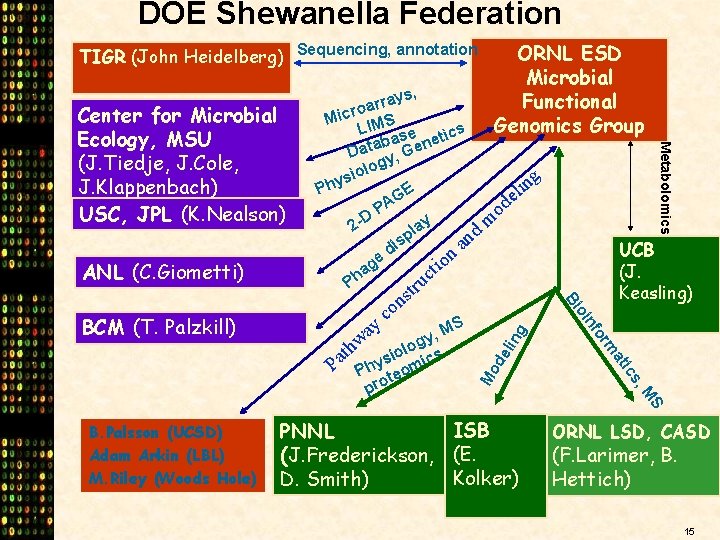
DOE Shewanella Federation TIGR (John Heidelberg) Sequencing, annotation g lin S M s, ic at rm fo S y, M g th siolo ics a P Phy eom t pro n oi y a w UCB (J. Keasling) Bi BCM (T. Palzkill) s n o c Mo de ANL (C. Giometti) ys, a r r oa Micr IMS L se netics a b Data y, Ge g olo i g s n i Phy GE el A d P o D y m 2 a d pl n s i a d n e o ti ag c h P tru Metabolomics Center for Microbial Ecology, MSU (J. Tiedje, J. Cole, J. Klappenbach) USC, JPL (K. Nealson) ORNL ESD Microbial Functional Genomics Group B. Palsson (UCSD) Adam Arkin (LBL) M. Riley (Woods Hole) ISB PNNL (J. Frederickson, (E. Kolker) D. Smith) ORNL LSD, CASD (F. Larimer, B. Hettich) 15

Large Genomes To Life Project: $38 M for 5 years Rapid Deduction of Stress Response Pathways in Metal/Radionuclide Reducing Bacteria Stress responses on: Desulfovibrio vulgaris Shewanella oneidensis Geobacter metallireducens National Laboratories Universities Private Organizations UC Berkeley U Washington U Missouri (Consultant) 16
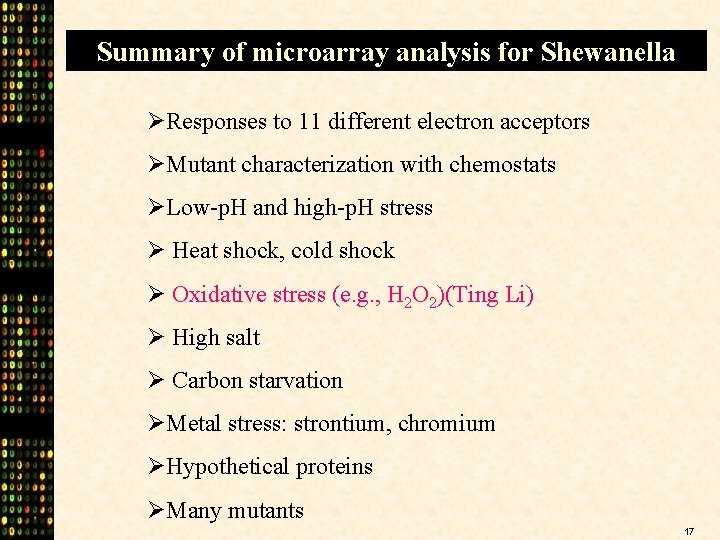
Summary of microarray analysis for Shewanella ØResponses to 11 different electron acceptors ØMutant characterization with chemostats ØLow-p. H and high-p. H stress Ø Heat shock, cold shock Ø Oxidative stress (e. g. , H 2 O 2)(Ting Li) Ø High salt Ø Carbon starvation ØMetal stress: strontium, chromium ØHypothetical proteins ØMany mutants 17
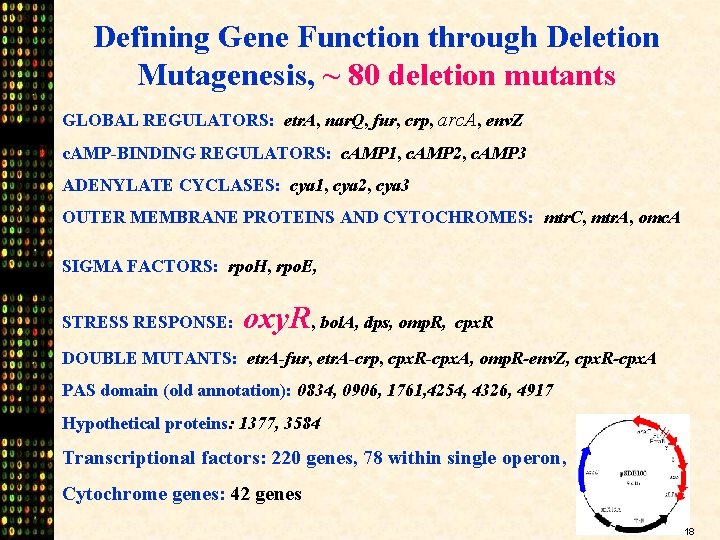
Defining Gene Function through Deletion Mutagenesis, ~ 80 deletion mutants GLOBAL REGULATORS: etr. A, nar. Q, fur, crp, arc. A, env. Z c. AMP-BINDING REGULATORS: c. AMP 1, c. AMP 2, c. AMP 3 ADENYLATE CYCLASES: cya 1, cya 2, cya 3 OUTER MEMBRANE PROTEINS AND CYTOCHROMES: mtr. C, mtr. A, omc. A SIGMA FACTORS: rpo. H, rpo. E, STRESS RESPONSE: oxy. R, bol. A, dps, omp. R, cpx. R DOUBLE MUTANTS: etr. A-fur, etr. A-crp, cpx. R-cpx. A, omp. R-env. Z, cpx. R-cpx. A PAS domain (old annotation): 0834, 0906, 1761, 4254, 4326, 4917 Hypothetical proteins: 1377, 3584 Transcriptional factors: 220 genes, 78 within single operon, Cytochrome genes: 42 genes 18
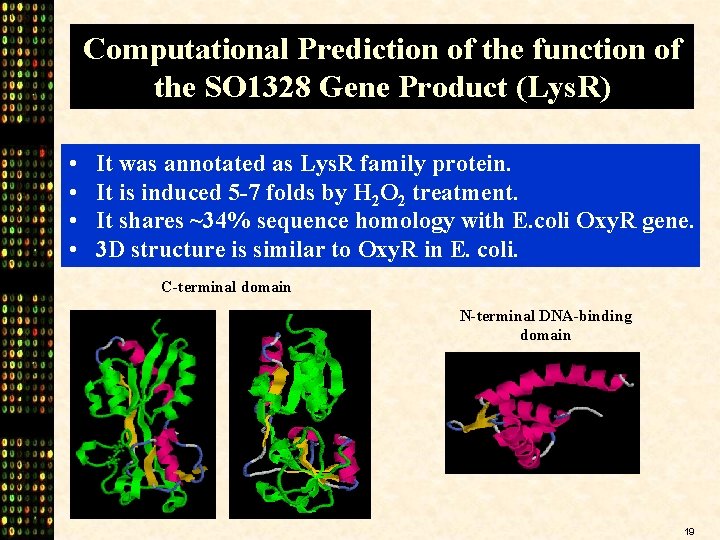
Computational Prediction of the function of the SO 1328 Gene Product (Lys. R) • • It was annotated as Lys. R family protein. It is induced 5 -7 folds by H 2 O 2 treatment. It shares ~34% sequence homology with E. coli Oxy. R gene. 3 D structure is similar to Oxy. R in E. coli. C-terminal domain N-terminal DNA-binding domain 19
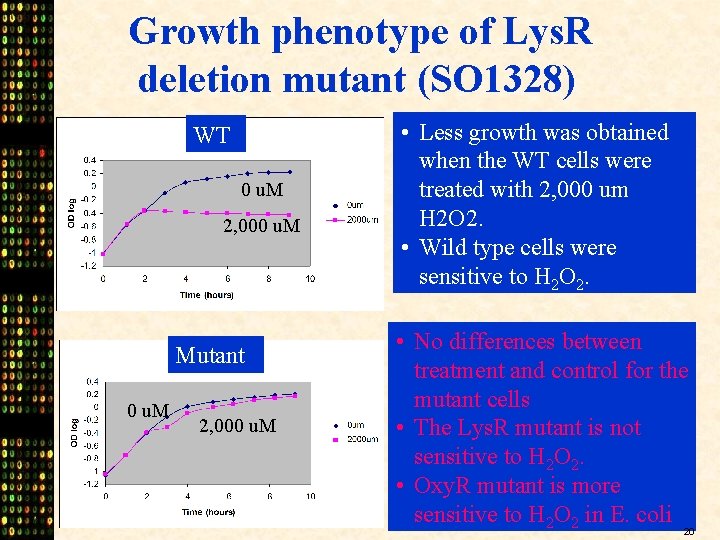
Growth phenotype of Lys. R deletion mutant (SO 1328) WT 0 u. M 2, 000 u. M Mutant 0 u. M 2, 000 u. M • Less growth was obtained when the WT cells were treated with 2, 000 um H 2 O 2. • Wild type cells were sensitive to H 2 O 2. • No differences between treatment and control for the mutant cells • The Lys. R mutant is not sensitive to H 2 O 2. • Oxy. R mutant is more sensitive to H 2 O 2 in E. coli 20
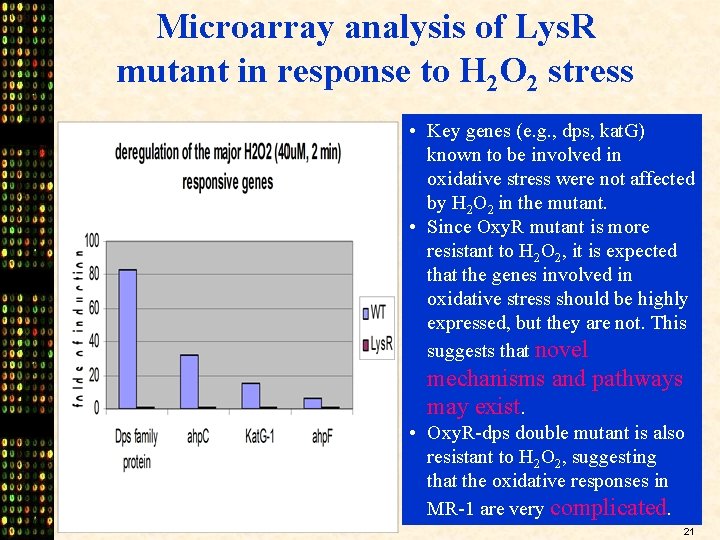
Microarray analysis of Lys. R mutant in response to H 2 O 2 stress • Key genes (e. g. , dps, kat. G) known to be involved in oxidative stress were not affected by H 2 O 2 in the mutant. • Since Oxy. R mutant is more resistant to H 2 O 2, it is expected that the genes involved in oxidative stress should be highly expressed, but they are not. This suggests that novel mechanisms and pathways may exist. • Oxy. R-dps double mutant is also resistant to H 2 O 2, suggesting that the oxidative responses in MR-1 are very complicated. 21
Proteomics Tools for studying proteomics Ø 2 -Dimentional gel electrophoresis ØMass spectrometry ØPhage-display ØYeast two hybrid system ØProtein arrays ØStructural determination: X-rays, NMR 22
Using phage-display to study protein interactions and regulations Gateway cloning vector Phage display • First key step: cloning all genes into universal vector. • The cloning systems were optimized. • All primers were synthesized. • 3, 853 genes were cloned. • Sequenced 50 clones, no errors were found. 23
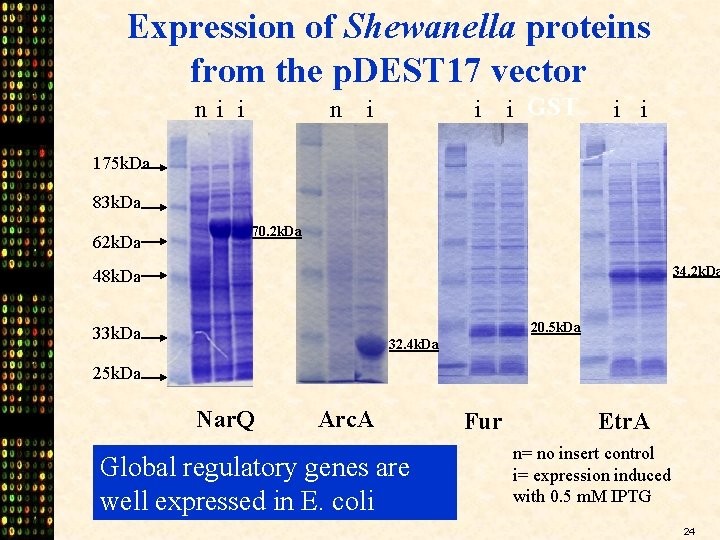
Expression of Shewanella proteins from the p. DEST 17 vector ni i n i i i GST i i 175 k. Da 83 k. Da 62 k. Da 70. 2 k. Da 34. 2 k. Da 48 k. Da 20. 5 k. Da 33 k. Da 32. 4 k. Da 25 k. Da Nar. Q Arc. A Global regulatory genes are well expressed in E. coli Fur Etr. A n= no insert control i= expression induced with 0. 5 m. M IPTG 24
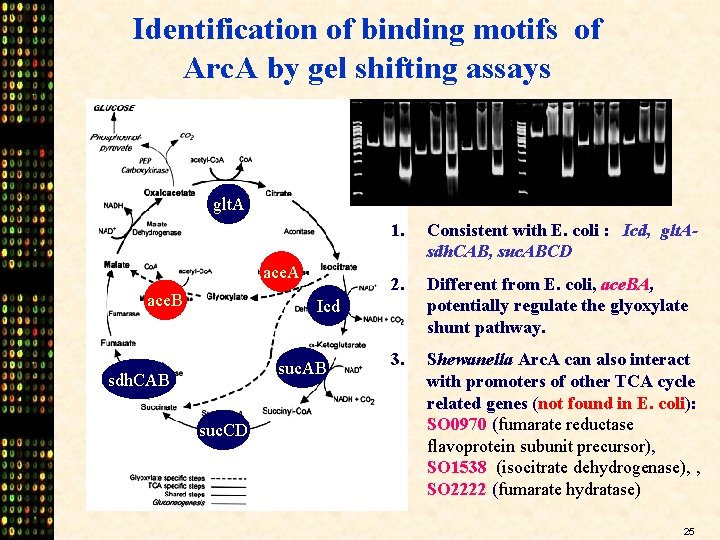
Identification of binding motifs of Arc. A by gel shifting assays glt. A ace. B 1. Consistent with E. coli : Icd, glt. Asdh. CAB, suc. ABCD 2. Different from E. coli, ace. BA, potentially regulate the glyoxylate shunt pathway. 3. Shewanella Arc. A can also interact with promoters of other TCA cycle related genes (not found in E. coli): SO 0970 (fumarate reductase flavoprotein subunit precursor), SO 1538 (isocitrate dehydrogenase), , SO 2222 (fumarate hydratase) Icd suc. AB sdh. CAB suc. CD 25
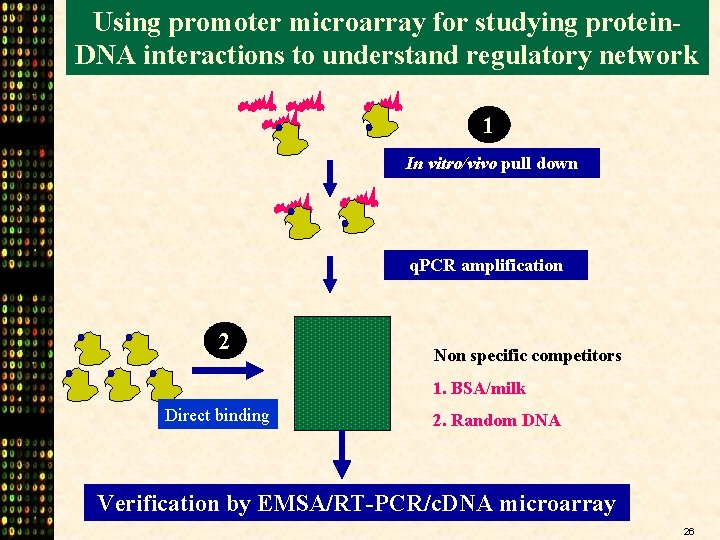
Using promoter microarray for studying protein. DNA interactions to understand regulatory network 1 In vitro/vivo pull down q. PCR amplification 2 Non specific competitors 1. BSA/milk Direct binding 2. Random DNA Verification by EMSA/RT-PCR/c. DNA microarray 26

Challenges in protein arrays · Antibodies are commonly used as probes in protein arrays · Two big challenges: - Loss of activity: The big challenge for antibody arrays is the loss of activity of antibody because the active binding site may bind to slide surface through chemical bonding, and thus the active site may not be available to the antigen. - Cross reactivity: Specificity is also a big issue for antibody protein arrays. . 27
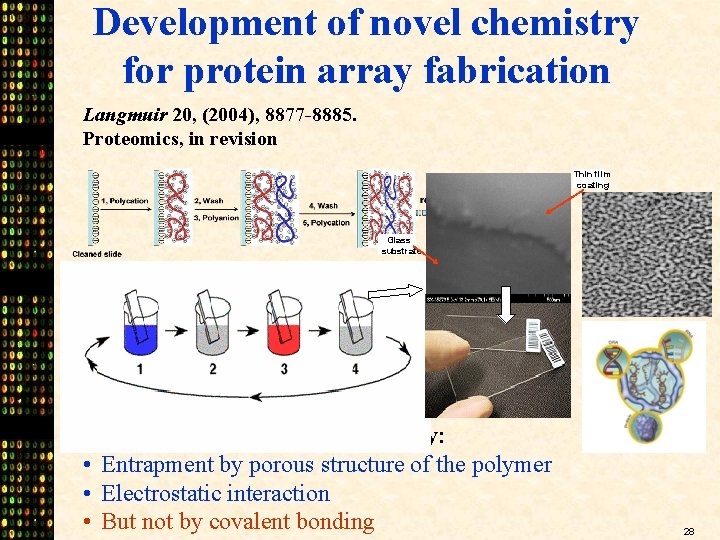
Development of novel chemistry for protein array fabrication Langmuir 20, (2004), 8877 -8885. Proteomics, in revision Thin film coating Glass substrate Proteins are affixed on the slide by: • Entrapment by porous structure of the polymer • Electrostatic interaction • But not by covalent bonding 28
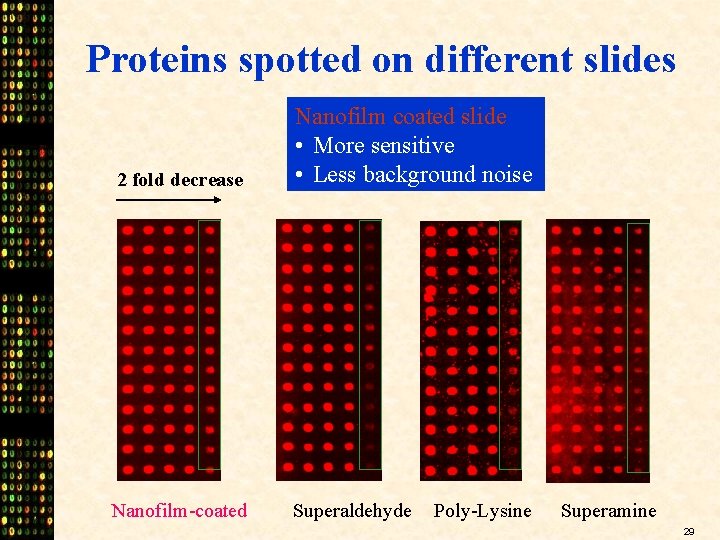
Proteins spotted on different slides 2 fold decrease Nanofilm coated slide • More sensitive • Less background noise Nanofilm-coated Superaldehyde Poly-Lysine Superamine 29
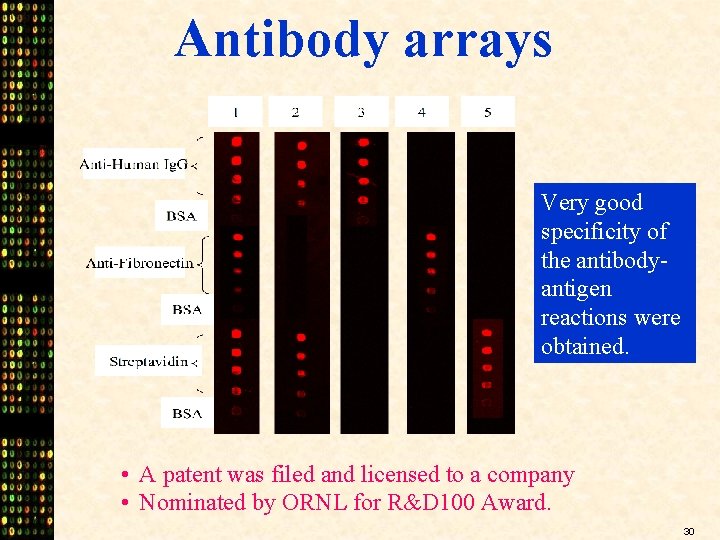
Antibody arrays Very good specificity of the antibodyantigen reactions were obtained. • A patent was filed and licensed to a company • Nominated by ORNL for R&D 100 Award. 30
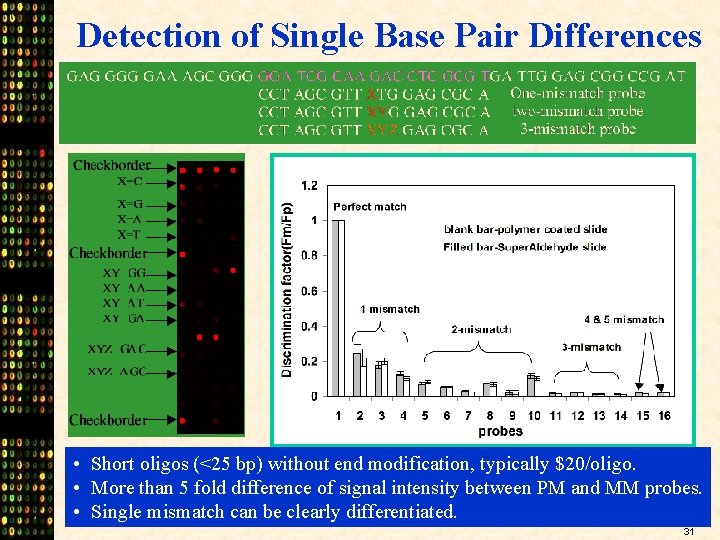
Detection of Single Base Pair Differences • Short oligos (<25 bp) without end modification, typically $20/oligo. • More than 5 fold difference of signal intensity between PM and MM probes. • Single mismatch can be clearly differentiated. 31
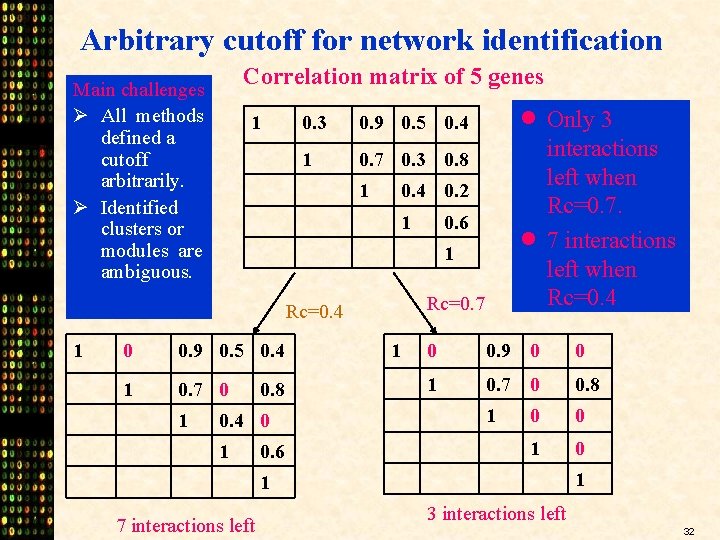
Arbitrary cutoff for network identification Correlation matrix of 5 genes Main challenges Ø All methods defined a cutoff arbitrarily. Ø Identified clusters or modules are ambiguous. 1 0. 3 0. 9 0. 5 0. 4 1 0. 7 0. 3 0. 8 1 0. 4 0. 2 1 0 0. 9 0. 5 0. 4 1 0. 7 0 1 0. 6 · 1 Rc=0. 7 Rc=0. 4 1 · Only 3 0. 8 0. 4 0 1 0. 6 1 interactions left when Rc=0. 7. 7 interactions left when Rc=0. 4 0 0. 9 0 0 1 0. 7 0 0. 8 1 0 0 1 1 7 interactions left 32
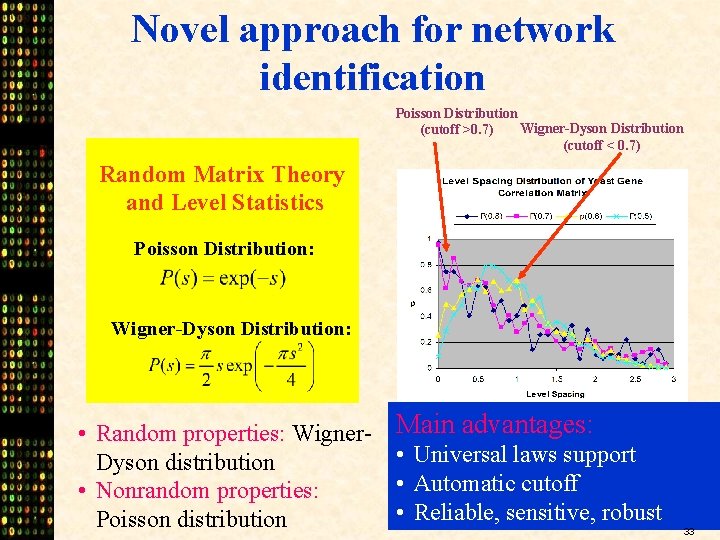
Novel approach for network identification Poisson Distribution Wigner-Dyson Distribution (cutoff >0. 7) (cutoff < 0. 7) Random Matrix Theory and Level Statistics Poisson Distribution: Wigner-Dyson Distribution: • Random properties: Wigner. Dyson distribution • Nonrandom properties: Poisson distribution Main advantages: • Universal laws support • Automatic cutoff • Reliable, sensitive, robust 33
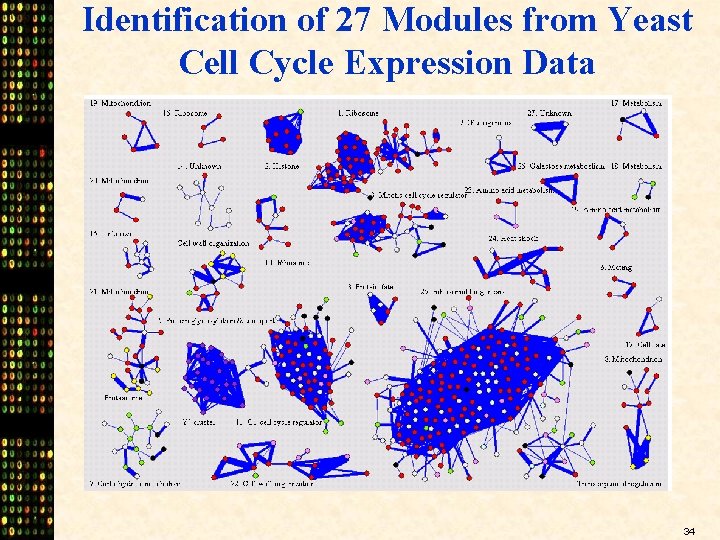
Identification of 27 Modules from Yeast Cell Cycle Expression Data 34
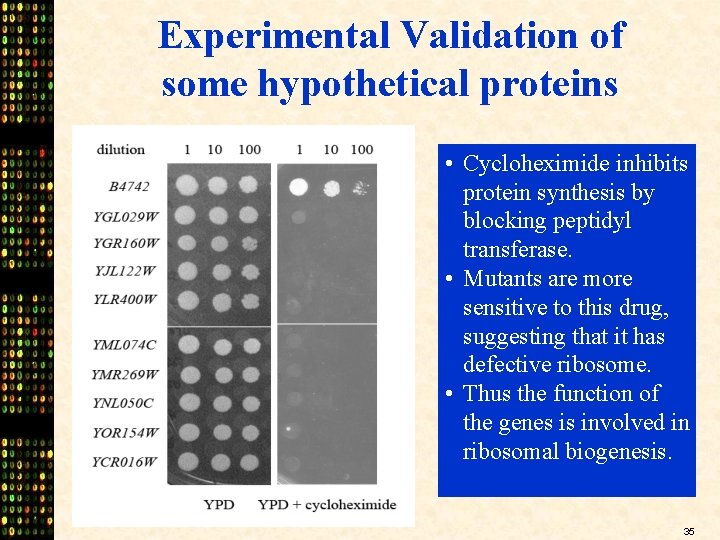
Experimental Validation of some hypothetical proteins • Cycloheximide inhibits protein synthesis by blocking peptidyl transferase. • Mutants are more sensitive to this drug, suggesting that it has defective ribosome. • Thus the function of the genes is involved in ribosomal biogenesis. 35
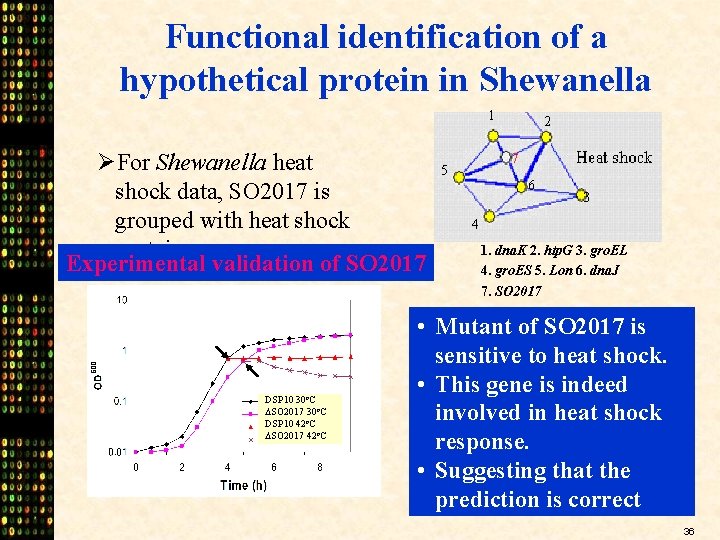
Functional identification of a hypothetical protein in Shewanella 1 ØFor Shewanella heat shock data, SO 2017 is grouped with heat shock proteins. Experimental validation of SO 2017 DSP 10 30 o. C SO 2017 30 o. C DSP 10 42 o. C SO 2017 42 o. C 2 7 5 6 3 4 1. dna. K 2. htp. G 3. gro. EL 4. gro. ES 5. Lon 6. dna. J 7. SO 2017 • Mutant of SO 2017 is sensitive to heat shock. • This gene is indeed involved in heat shock response. • Suggesting that the prediction is correct 36
Pioneering advances in microarray-based technologies to address challenges in microbial community genomics · Challenges: - Specificity: Environmental sequence divergences. Sensitivity: Low biomass. Quantification: Existence of contaminants: Humic materials, organic contaminants, metals and radionuclides. · Solutions - Developing different types of microarrays and novel chemistry to address different levels of specificity. - Developing novel signal amplification strategy to increase sensitivity - Optimizing microarray protocols for reliable quantification. 37

Summary of 50 mer-based FGAs for environmental studies Oligonucleotide probe size: 50 bp Tiquia et al. 2004. Bio. Techniques 36, 664 -675 Rhee et al. 2004, AEM 70: 4303 -4317 • • • Nitrogen cycling: 302 Sulfate reduction: 204 Carbon cycling: 566 Phosphorus utilization: 79 Organic contaminant degradation: 770 Metal resistance and oxidation: 85 • Total: 2, 006 probes • All probes are < 88% similarity 38
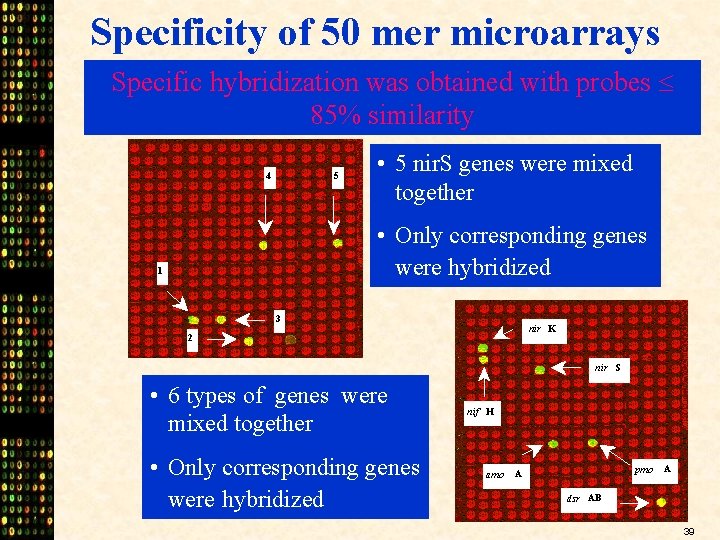
Specificity of 50 mer microarrays Specific hybridization was obtained with probes 85% similarity 4 5 • 5 nir. S genes were mixed together • Only corresponding genes were hybridized 1 3 nir K 2 nir S • 6 types of genes were mixed together • Only corresponding genes were hybridized nif H amo pmo A A dsr AB 39
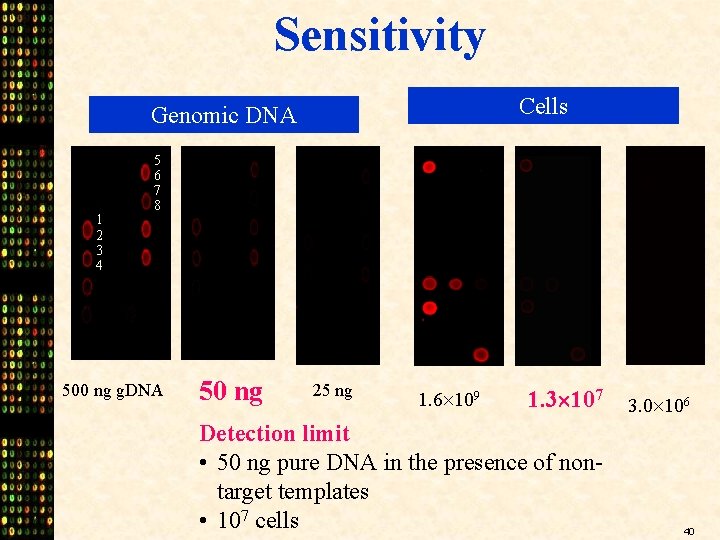
Sensitivity Cells Genomic DNA 1 2 3 4 5 6 7 8 500 ng g. DNA 50 ng 25 ng 1. 6 109 1. 3 107 Detection limit • 50 ng pure DNA in the presence of nontarget templates • 107 cells 3. 0 106 40
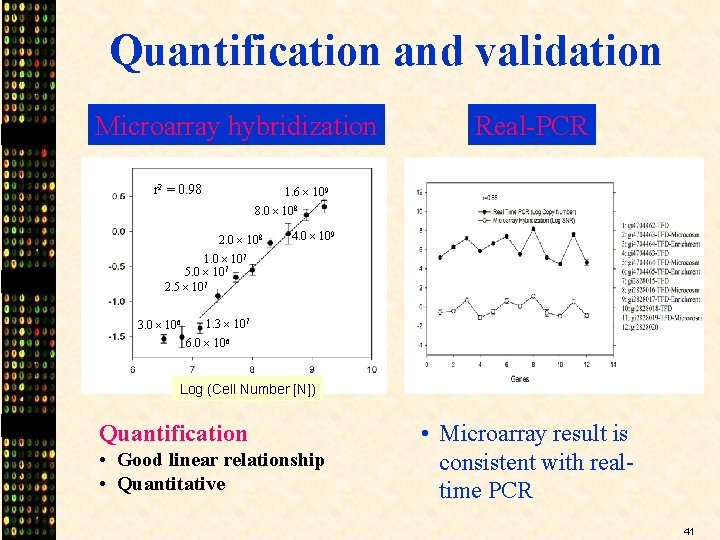
Quantification and validation Microarray hybridization r 2 = 0. 98 Real-PCR 1. 6 109 8. 0 108 2. 0 108 4. 0 109 1. 0 107 5. 0 107 2. 5 107 3. 0 106 1. 3 107 6. 0 106 Log (Cell Number [N]) Quantification • Good linear relationship • Quantitative • Microarray result is consistent with realtime PCR 41
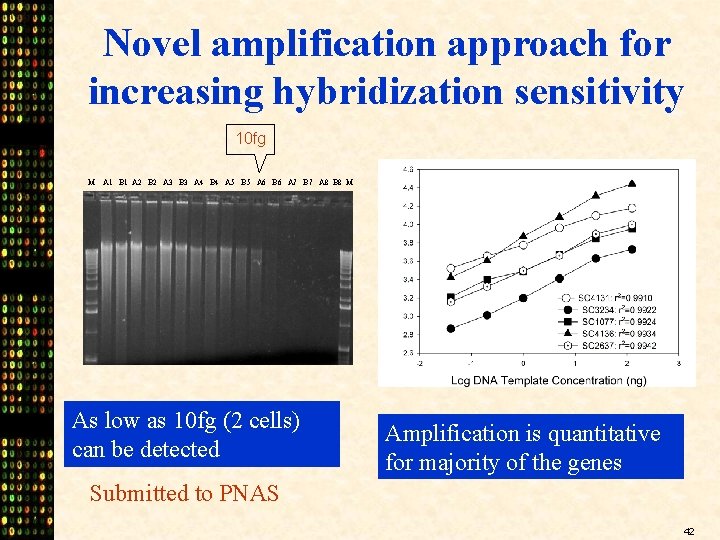
Novel amplification approach for increasing hybridization sensitivity 10 fg M A 1 B 1 A 2 B 2 A 3 B 3 A 4 B 4 A 5 B 5 A 6 B 6 A 7 B 7 A 8 B 8 M As low as 10 fg (2 cells) can be detected Amplification is quantitative for majority of the genes Submitted to PNAS 42
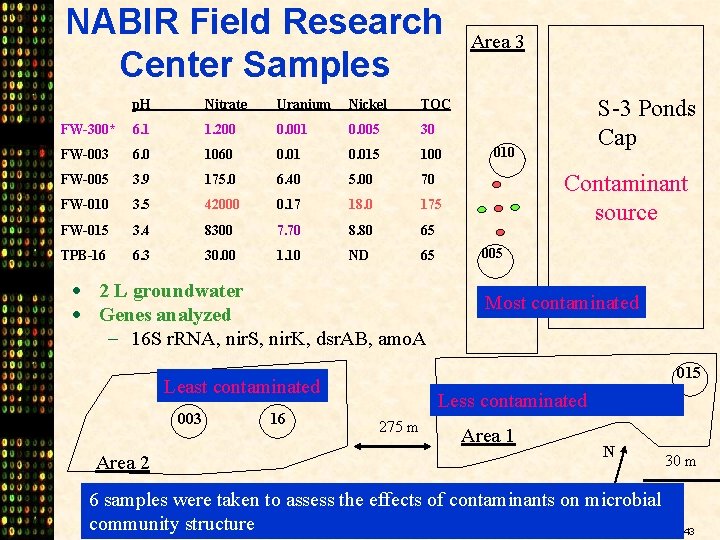
NABIR Field Research Center Samples p. H Nitrate Uranium Nickel TOC FW-300* 6. 1 1. 200 0. 001 0. 005 30 FW-003 6. 0 1060 0. 015 100 FW-005 3. 9 175. 0 6. 40 5. 00 70 FW-010 3. 5 42000 0. 17 18. 0 175 FW-015 3. 4 8300 7. 70 8. 80 65 TPB-16 6. 3 30. 00 1. 10 ND 65 · 2 L groundwater · Genes analyzed - 16 S r. RNA, nir. S, nir. K, dsr. AB, amo. A Area 2 16 S-3 Ponds Cap 010 Contaminant source 005 Most contaminated 015 Least contaminated 003 Area 3 Less contaminated 275 m Area 1 N 30 m 6 samples were taken to assess the effects of contaminants on microbial community structure 43
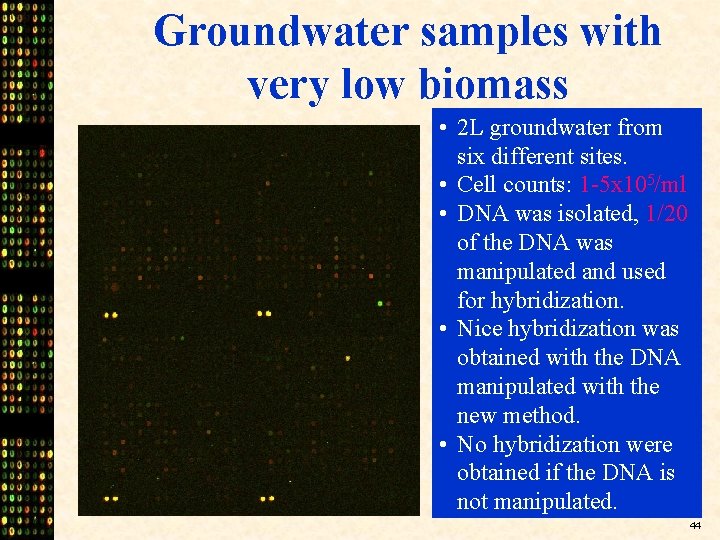
Groundwater samples with very low biomass • 2 L groundwater from six different sites. • Cell counts: 1 -5 x 105/ml • DNA was isolated, 1/20 of the DNA was manipulated and used for hybridization. • Nice hybridization was obtained with the DNA manipulated with the new method. • No hybridization were obtained if the DNA is not manipulated. 44
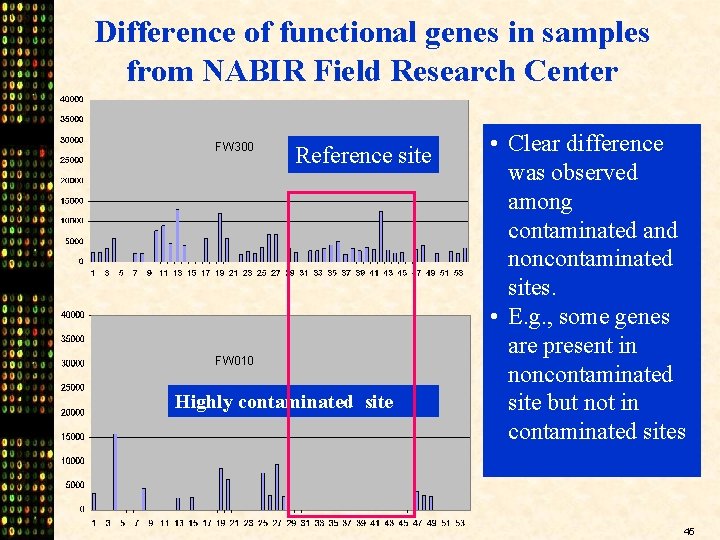
Difference of functional genes in samples from NABIR Field Research Center FW 300 Reference site FW 010 Highly contaminated site • Clear difference was observed among contaminated and noncontaminated sites. • E. g. , some genes are present in noncontaminated site but not in contaminated sites 45
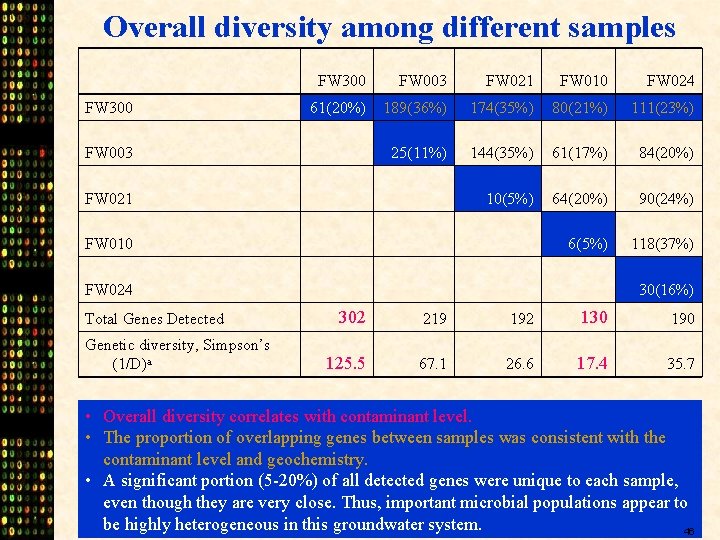
Overall diversity among different samples FW 300 FW 003 FW 021 FW 010 FW 024 61(20%) 189(36%) 174(35%) 80(21%) 111(23%) 25(11%) 144(35%) 61(17%) 84(20%) 10(5%) 64(20%) 90(24%) 6(5%) 118(37%) FW 003 FW 021 FW 010 FW 024 Total Genes Detected Genetic diversity, Simpson’s (1/D)a 30(16%) 302 219 192 130 190 125. 5 67. 1 26. 6 17. 4 35. 7 • Overall diversity correlates with contaminant level. • The proportion of overlapping genes between samples was consistent with the contaminant level and geochemistry. • A significant portion (5 -20%) of all detected genes were unique to each sample, even though they are very close. Thus, important microbial populations appear to be highly heterogeneous in this groundwater system. 46
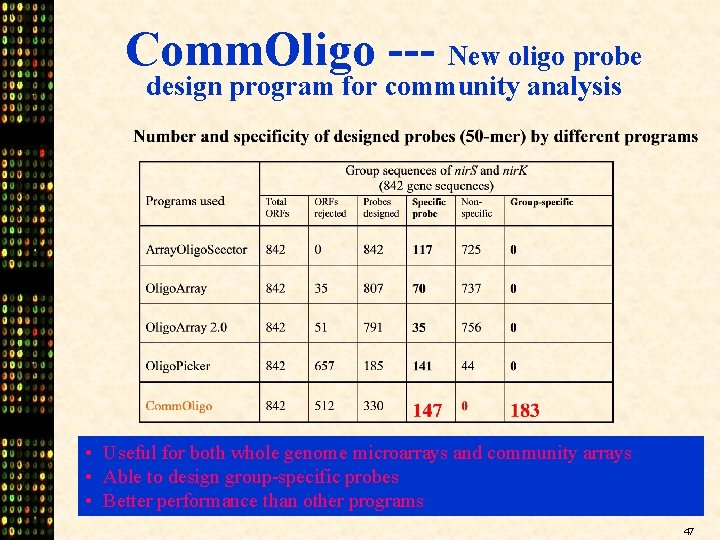
Comm. Oligo --- New oligo probe design program for community analysis • Useful for both whole genome microarrays and community arrays • Able to design group-specific probes • Better performance than other programs 47
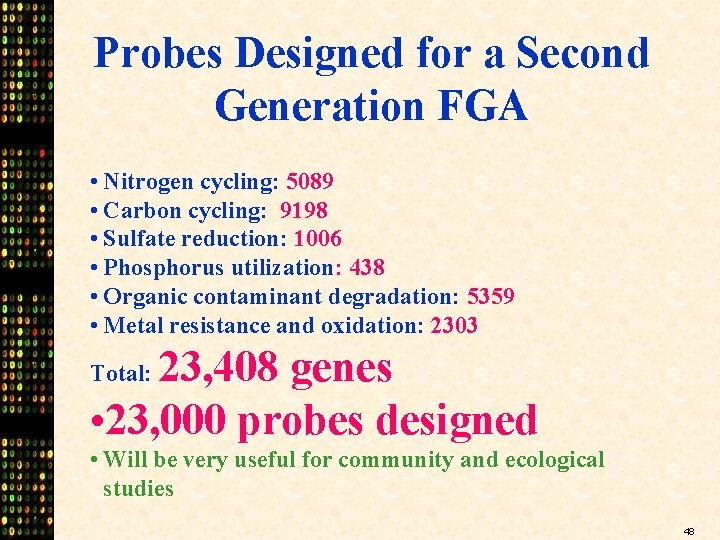
Probes Designed for a Second Generation FGA • Nitrogen cycling: 5089 • Carbon cycling: 9198 • Sulfate reduction: 1006 • Phosphorus utilization: 438 • Organic contaminant degradation: 5359 • Metal resistance and oxidation: 2303 23, 408 genes • 23, 000 probes designed Total: • Will be very useful for community and ecological studies 48

Community Genomics Grand challenges • Extremely high diversity, 5000 species/g soil • 99% of the microbial species are uncultured 49
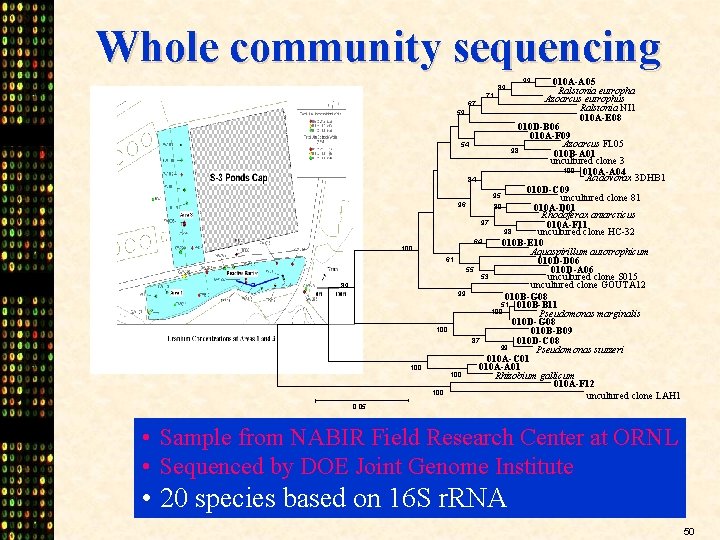
Whole community sequencing 010 A-A 05 Ralstonia eutropha Azoarcus eutrophus 67 Ralstonia NI 1 59 010 A-E 08 010 D-B 06 010 A-F 09 54 Azoarcus FL 05 98 010 B-A 01 uncultured clone 3 100 010 A-A 04 Acidovorax 3 DHB 1 84 010 D-C 09 95 uncultured clone 81 96 80 010 A-D 01 Rhodoferax antarcticus 97 010 A-F 11 98 uncultured clone HC-32 64 010 B-E 10 Aquaspirillum autotrophicum 61 010 D-D 06 55 010 D-A 06 53 uncultured clone S 015 uncultured clone GOUTA 12 99 010 B-G 08 51 010 B-B 11 100 Pseudomonas marginalis 010 D-G 08 100 010 B-B 09 87 010 D-C 08 99 Pseudomonas stutzeri 010 A-C 01 010 A-A 01 100 Rhizobium gallicum 010 A-F 12 100 uncultured clone LAH 1 89 99 71 100 89 100 0. 05 • Sample from NABIR Field Research Center at ORNL • Sequenced by DOE Joint Genome Institute • 20 species based on 16 S r. RNA 50
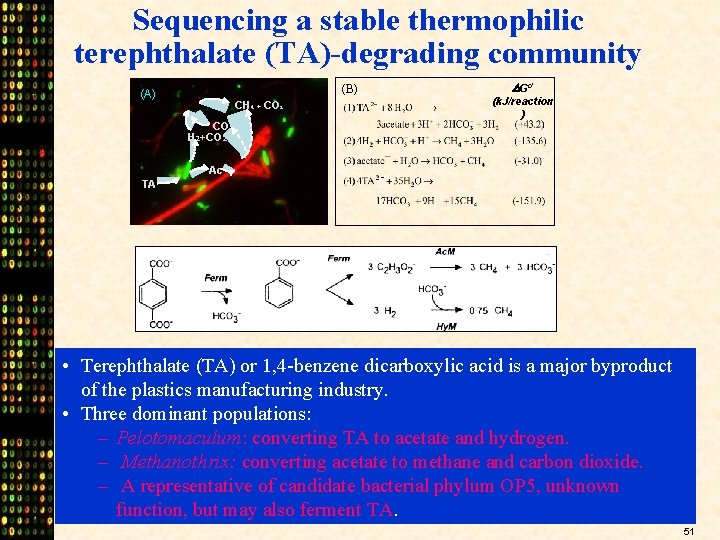
Sequencing a stable thermophilic terephthalate (TA)-degrading community (B) (A) CH 4 + CO 2 Go’ (k. J/reaction ) CO 2 H 2+CO 2 Ac TA • Terephthalate (TA) or 1, 4 -benzene dicarboxylic acid is a major byproduct of the plastics manufacturing industry. • Three dominant populations: – Pelotomaculum: converting TA to acetate and hydrogen. – Methanothrix: converting acetate to methane and carbon dioxide. – A representative of candidate bacterial phylum OP 5, unknown function, but may also ferment TA. 51
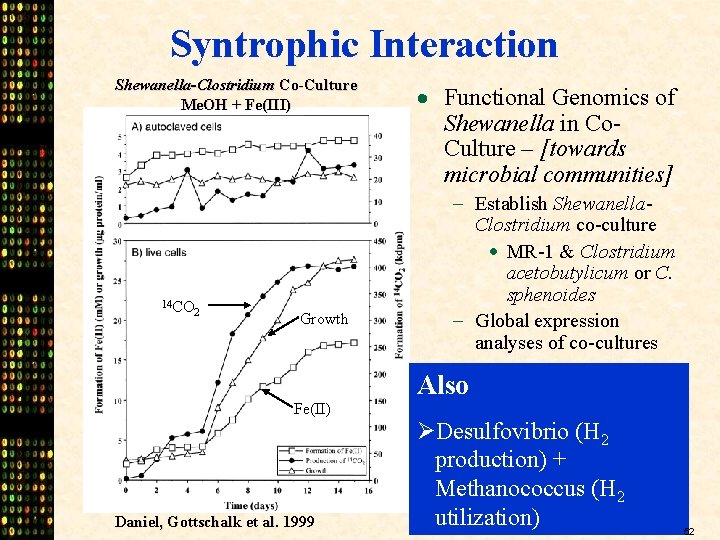
Syntrophic Interaction Shewanella-Clostridium Co-Culture Me. OH + Fe(III) 14 CO 2 Growth Fe(II) Daniel, Gottschalk et al. 1999 · Functional Genomics of Shewanella in Co. Culture – [towards microbial communities] - Establish Shewanella. Clostridium co-culture · MR-1 & Clostridium acetobutylicum or C. sphenoides - Global expression analyses of co-cultures Also ØDesulfovibrio (H 2 production) + Methanococcus (H 2 utilization) 52
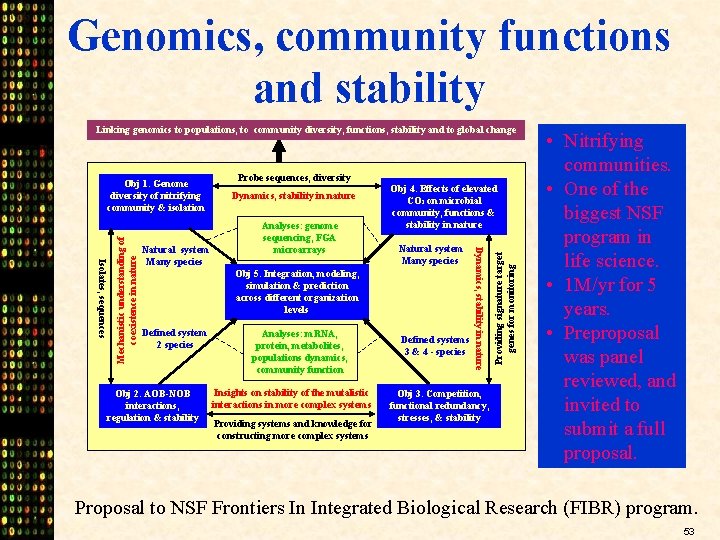
Genomics, community functions and stability Linking genomics to populations, to community diversity, functions, stability and to global change Dynamics, stability in nature Analyses: genome sequencing, FGA microarrays Obj 4. Effects of elevated CO 2 on microbial community, functions & stability in nature Natural system Many species Obj 5. Integration, modeling, simulation & prediction across different organization levels Defined system 2 species Obj 2. AOB-NOB interactions, regulation & stability Analyses: m. RNA, protein, metabolites, populations dynamics, community function Insights on stability of the mutalistic interactions in more complex systems Providing systems and knowledge for constructing more complex systems Defined systems 3 & 4 - species Obj 3. Competition, functional redundancy, stresses, & stability Providing signature target genes for monitoring Natural system Many species Probe sequences, diversity Dynamics, stability in nature Isolates, sequences Mechanistic understanding of coexistence in nature Obj 1. Genome diversity of nitrifying community & isolation • Nitrifying communities. • One of the biggest NSF program in life science. • 1 M/yr for 5 years. • Preproposal was panel reviewed, and invited to submit a full proposal. Proposal to NSF Frontiers In Integrated Biological Research (FIBR) program. 53
Predictive Microbial Ecology · Qualitative microbial ecology: Due to the difficulty in obtaining experimental data, microbial ecology is qualitative, but not quantitative. · Opportunity for quantitative microbial science: With availability of genomic technologies, microbial ecology is no longer limited by the deficiency of experimental data. · Challenges: Modeling, simulation and prediction - A big mathematical challenges: dimensionality problem. The sample number is less than the gene number. · Possible solution: System ecology + Genomics An example of the conceptual integration scheme i. Modeling microarray data at individual gene level ii. Modeling interactions between functional gene groups or gilds. 54
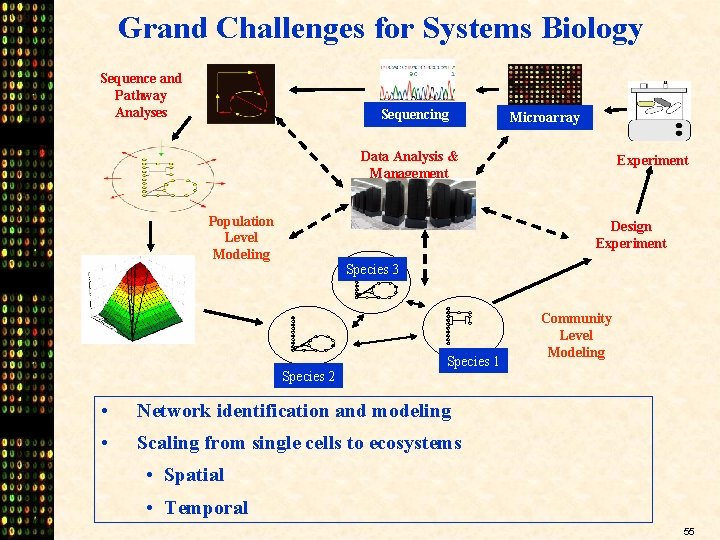
Grand Challenges for Systems Biology Sequence and Pathway Analyses Sequencing Microarray Data Analysis & Management Population Level Modeling Experiment Design Experiment Species 3 Species 2 Species 1 • Network identification and modeling • Scaling from single cells to ecosystems Community Level Modeling • Spatial • Temporal 55

First Book on Microbial Functional Genomics · Authors - Jizhong Zhou, Dorothea Thompson, Ying Xu, James M. Tiedje · · John Wiley & Sons, March 19, 2004 15 chapters, > 600 pages Rita Colwell, former NSF Director, wrote a forward To our knowledge, this is the first book in microbial functional genomics 56

Acknowledgement (1) • Department of Energy – Microbial Genome Program – Genomes To Life Program – NABIR Program – Ocean Margin Program – Carbon cycling programs • Oak Ridge National Laboratory – Laboratory Directed Research and Development 57

Microbial Genomics and Ecology Group at Environmental Sciences Division, ORNL 58
• ORNL – – – – – – – – Acknowledgement Zhili He Liyou Wu Dorothea Thompson Yongqing Liu Ting Li Matthew Fields Xuedan Liu Tingfen Yan Sung-Keun Rhee Song Chong Yunfeng Yang Jost Liebich Christopher Schadt Dawn Stanek Adam Leaphart Weimin Gao Terry Gentry Steve Brown Qiang He Feng Luo Crystal Mc. Alvin Susan Carroll Lisa Fagan Haichun Gao Hongbin Pan Xiufeng Wan Xichun Zhou Zamin Yang Jianxin Zhong Dong Yu Ying Xu • Michigan State University – – – James M. Tiedje James Cole Joel Klappenbach • USUHS – Mike Daly • USC – Ken Nealson • Argonne National Lab – Carol Giomettie • Univ of Iowa – Caroline Harwood • Oregon State Univ – Dan Arp • UC Berkeley – Jay Kneasling • Ohio State Univ – Bob Tabita • Univ of Missouri – Judy Wall • Bayler College – Tim Palzkill • SREL – Chuanlun Zhang • PNNL – – – Jim Frederickson Margie Romine Yuri Gorby Dick Smith Mary Lipton • LBL – – Terry Hazen Adam Arkin • Perkin Elmer – Xinyuan Li 59
- Slides: 59