Microarrays vs ultrasequencing Barbara Radovani Genomics Master of
Microarrays vs ultrasequencing Barbara Radovani Genomics Master of Advanced genetics, UAB
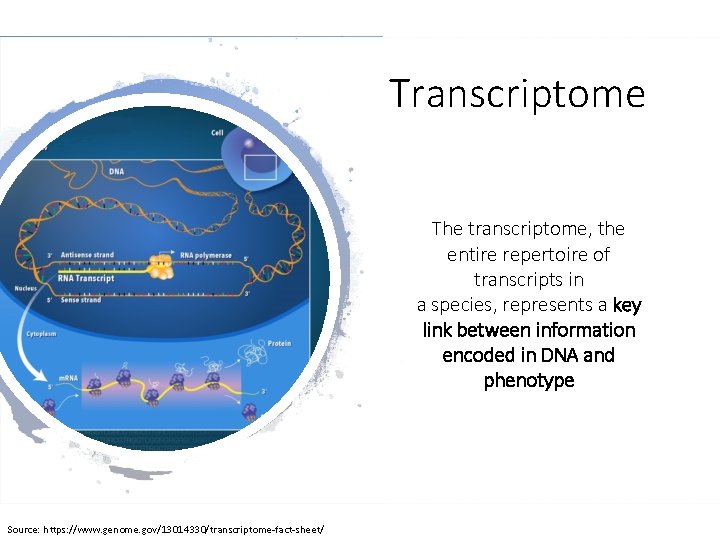
Transcriptome The transcriptome, the entire repertoire of transcripts in a species, represents a key link between information encoded in DNA and phenotype Source: https: //www. genome. gov/13014330/transcriptome-fact-sheet/
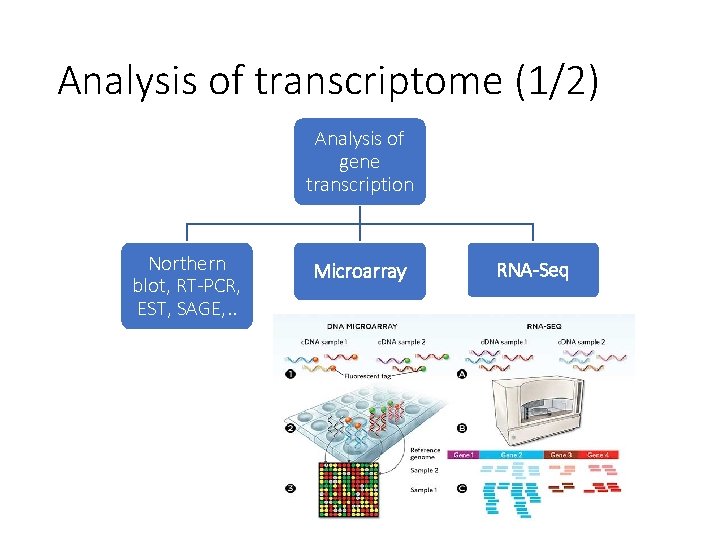
Analysis of transcriptome (1/2) Analysis of gene transcription Northern blot, RT-PCR, EST, SAGE, . . Microarray RNA-Seq
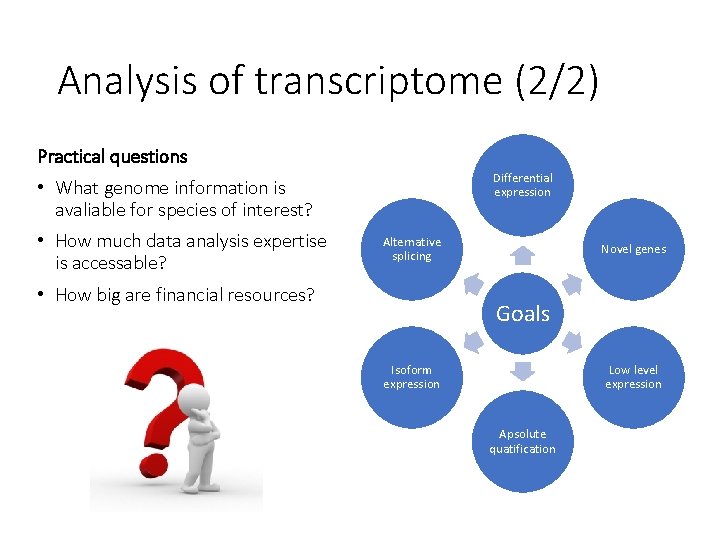
Analysis of transcriptome (2/2) Practical questions Differential expression • What genome information is avaliable for species of interest? • How much data analysis expertise is accessable? Alternative splicing • How big are financial resources? Novel genes Goals Isoform expression Low level expression Apsolute quatification
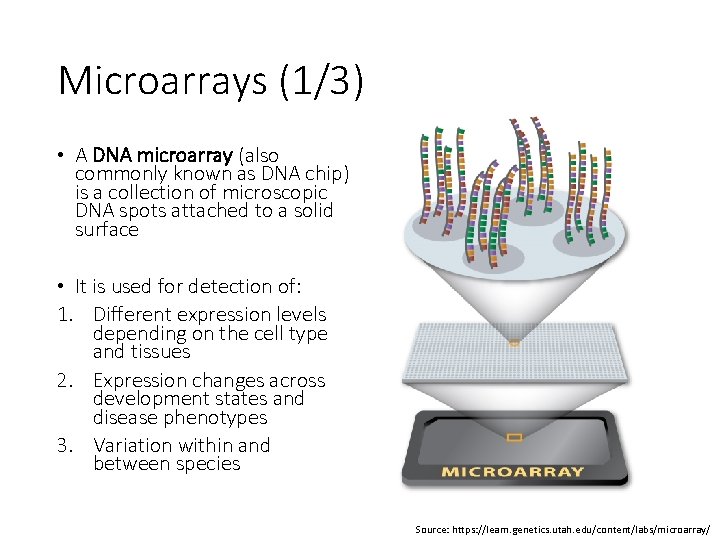
Microarrays (1/3) • A DNA microarray (also commonly known as DNA chip) is a collection of microscopic DNA spots attached to a solid surface • It is used for detection of: 1. Different expression levels depending on the cell type and tissues 2. Expression changes across development states and disease phenotypes 3. Variation within and between species Source: https: //learn. genetics. utah. edu/content/labs/microarray/
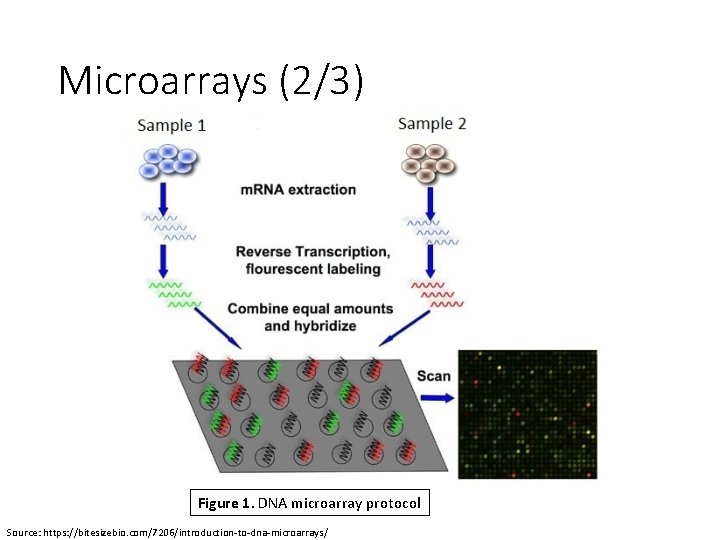
Microarrays (2/3) Figure 1. DNA microarray protocol Source: https: //bitesizebio. com/7206/introduction-to-dna-microarrays/
Microarrays (3/3) ADVANTAGES • Reliable method, standardized protocols over decades of use • Straightforward data analysis • Low cost DRAWBACKS • Dependant on prior sequence knowledge • Cannot detect structural variations • Cannot detect isoforms • Hybridization and sample labeling BIAS-es
RNA-Seq (1/3) • RNA-Seq is a powerful sequencing-based method that enables researchers to discover, profile, and quantify RNA transcripts across the entire transcriptome • DNA fragments are repeatedly sequenced in a very short time – deep sequencing increased sensitivity and accuracy http: //omegabioservices. com/index. php/next-gen-sequencing/rna-seq/
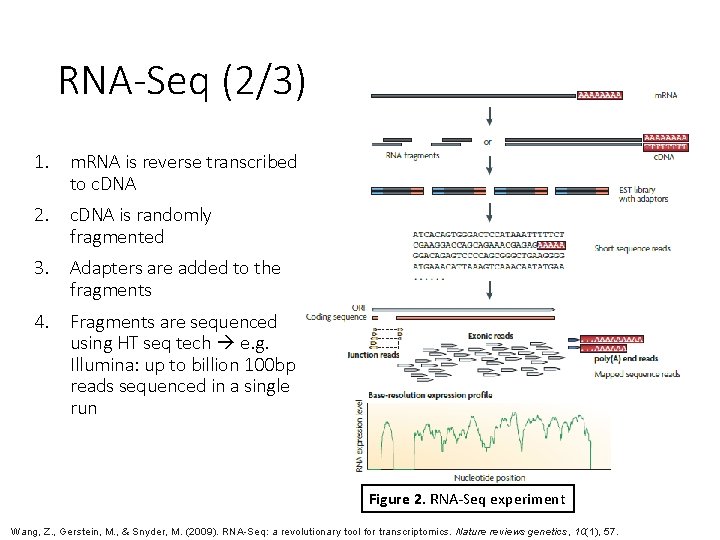
RNA-Seq (2/3) 1. m. RNA is reverse transcribed to c. DNA 2. c. DNA is randomly fragmented 3. Adapters are added to the fragments 4. Fragments are sequenced using HT seq tech e. g. Illumina: up to billion 100 bp reads sequenced in a single run Figure 2. RNA-Seq experiment Wang, Z. , Gerstein, M. , & Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews genetics, 10(1), 57.
RNA-Seq (3/3) ADVANTAGES • Not dependant on any prior sequence knowledge direct sequence alignment instead of hybridization • Alternative splicing can be detected • SNP identification • High dynamic range DRAWBACKS • Protocols still not fully optimised • Data storage is more challenging • Requires high power computing facillities and specialized personnel • More expensive than microarrays depends on genome complexity
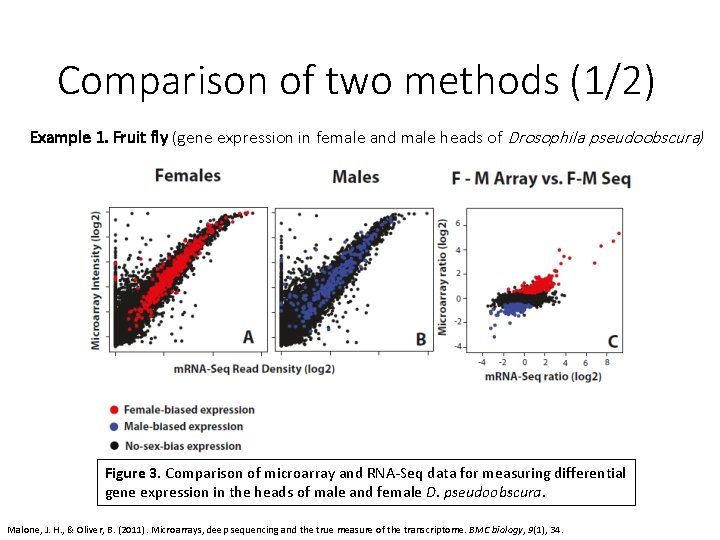
Comparison of two methods (1/2) Example 1. Fruit fly (gene expression in female and male heads of Drosophila pseudoobscura) Figure 3. Comparison of microarray and RNA-Seq data for measuring differential gene expression in the heads of male and female D. pseudoobscura. Malone, J. H. , & Oliver, B. (2011). Microarrays, deep sequencing and the true measure of the transcriptome. BMC biology, 9(1), 34.
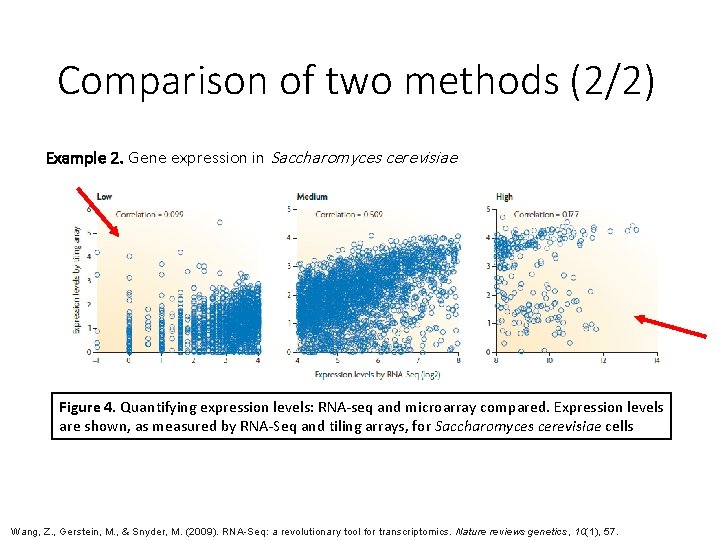
Comparison of two methods (2/2) Example 2. Gene expression in Saccharomyces cerevisiae Figure 4. Quantifying expression levels: RNA-seq and microarray compared. Expression levels are shown, as measured by RNA-Seq and tiling arrays, for Saccharomyces cerevisiae cells Wang, Z. , Gerstein, M. , & Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews genetics, 10(1), 57.
Conclusion • Both sequencing and hybridizing m. RNA to arrays are HT ways to profile the transcriptome and for problems that can be addressed by both, they show similar performance • Detecting genes with low expression is still a problem for both techniques, but there are some applications, such as transcript discovery and isoform identification, where RNA-Seq will be the better choice • As the cost of sequencing continues to fall, RNA-Seq is expected to replace microarrays for many applications that involve determining the structure and dynamics of the transcriptome
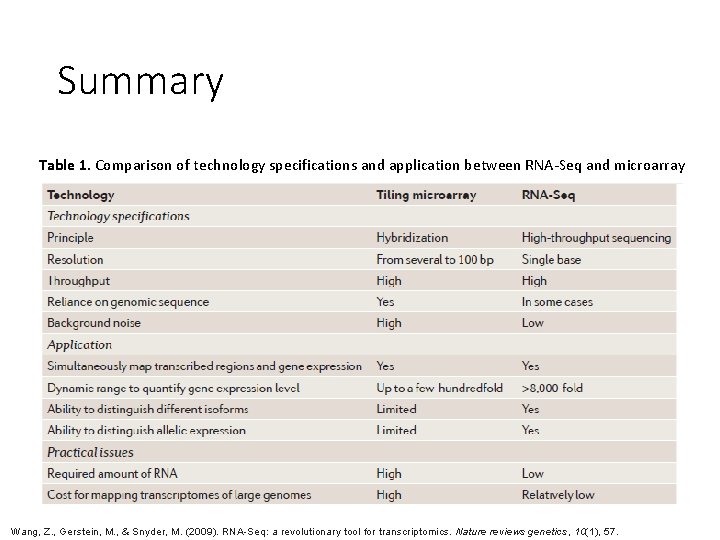
Summary Table 1. Comparison of technology specifications and application between RNA-Seq and microarray Wang, Z. , Gerstein, M. , & Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews genetics, 10(1), 57.

THANK YOU FOR YOUR ATTENTION!

References • Malone, J. H. , & Oliver, B. (2011). Microarrays, deep sequencing and the true measure of the transcriptome. BMC biology, 9(1), 34. • Source: Wang, Z. , Gerstein, M. , & Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews genetics, 10(1), 57. • Liu, S. , Lin, L. , Jiang, P. , Wang, D. , & Xing, Y. (2010). A comparison of RNASeq and high-density exon array for detecting differential gene expression between closely related species. Nucleic acids research, 39(2), 578 -588.
- Slides: 16