Microarray Data Anaysis and ICA Kim Hye Jin
- Slides: 15

Microarray Data Anaysis and ICA Kim, Hye. Jin Intelligent Multimedia Lab. Department of Computer Science and Engineering POSTECH, Korea seungjin@postech. ac. kr

Introduction n Microarray Data Analysis n Data Analysis Tools n n Supervised learning methods n Unsupervised learning methods ICA applied to Microarray Data n Time courses n n Different cell types n n Yeast cell cycle data B-cell lymphoma data Discussion

Microarray Data(1)

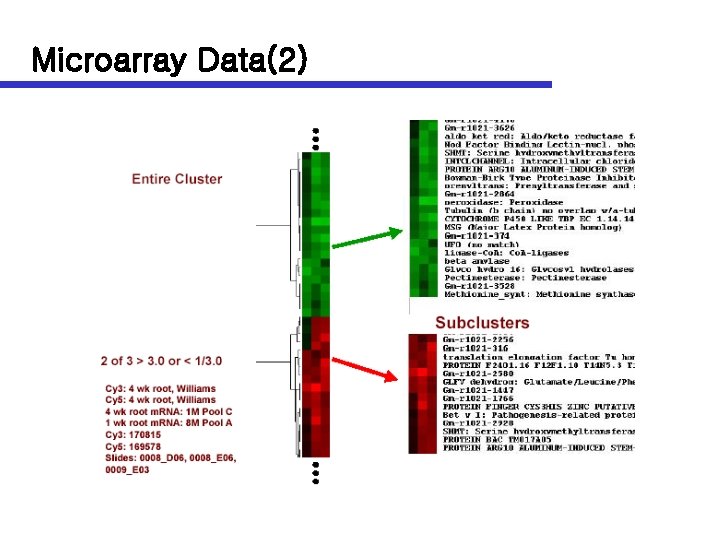
Microarray Data(2)

Data Analysis Tools n Supervised learning methods n n n Classification problem cases Partial Least Squares(PLS), Kernel PLS, Support Vector machine(SVM) Unsupervised learning methods n n Clustering problem cases Hierarchical clustering, Self-organized map(SOM), Singular value decomposition(SVD), plaid model, Principal component analysis (PCA) and Independent component analysis(ICA)

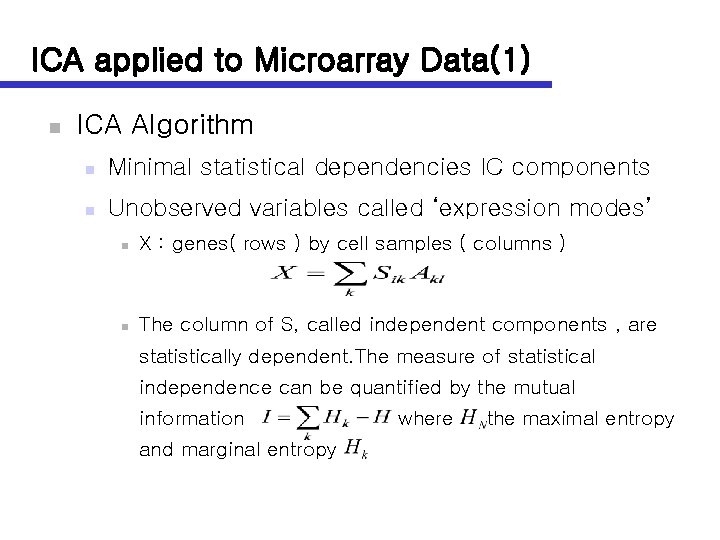
ICA applied to Microarray Data(1) n ICA Algorithm n Minimal statistical dependencies IC components n Unobserved variables called ‘expression modes’ n X : genes( rows ) by cell samples ( columns ) n The column of S, called independent components , are statistically dependent. The measure of statistical independence can be quantified by the mutual information and marginal entropy where the maximal entropy

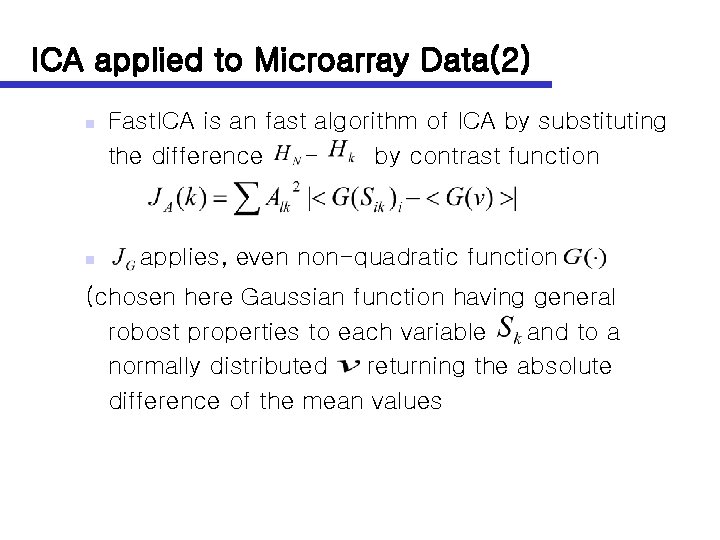
ICA applied to Microarray Data(2) n n Fast. ICA is an fast algorithm of ICA by substituting the difference by contrast function applies, even non-quadratic function (chosen here Gaussian function having general robost properties to each variable and to a normally distributed returning the absolute difference of the mean values

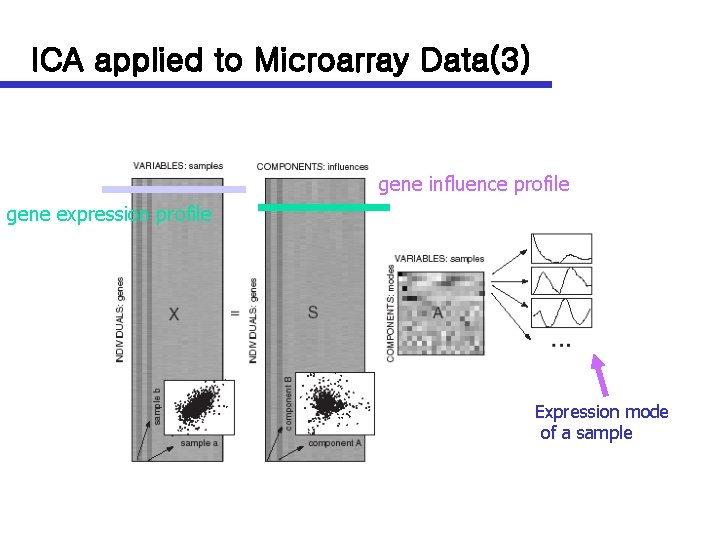
ICA applied to Microarray Data(3) gene influence profile gene expression profile Expression mode of a sample

ICA applied to Microarray Data(4) n Linear models of gene expression n Model assumptions n n Expression modes : the sample expression profiles are determined by a combination of hidden regulatory variables The genes responses to “expression mode” can be approximated by linear functions A linear model should reflect plausible properties of effective biological regulators ICA advantage n n Sensitive to modes whose influences on the genes follow ‘supergaussian’ distribution with large tails and a pronounced peak in the middle The reduce model ( Bussemaker et. al, 2001) is based on the occurrence of common motifs in the genes’ promoter sequences

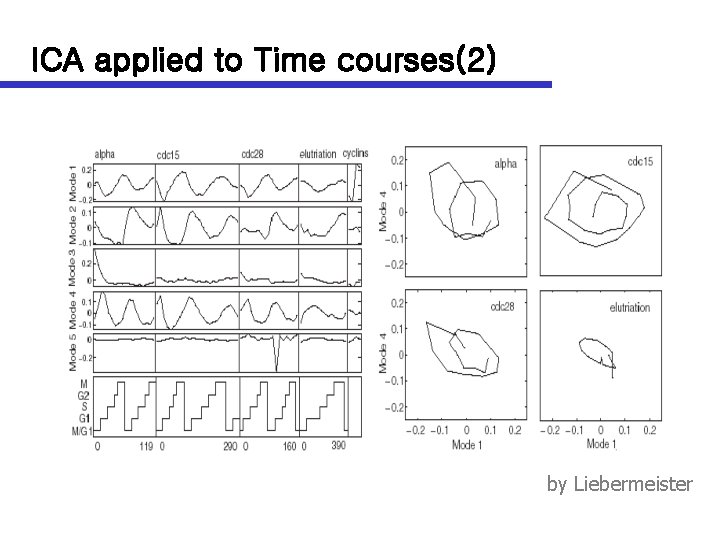
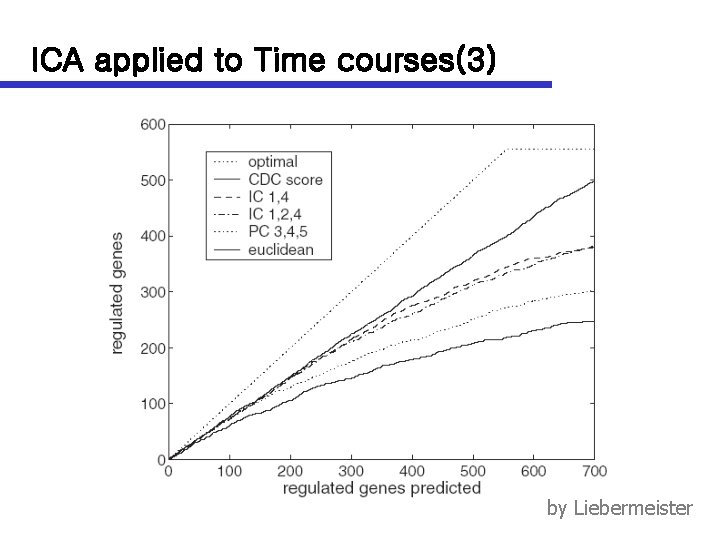
ICA applied to Time courses(1) n Time courses n Yeast cell cycle data n 77 by 6178 ORF expression ( Spellman et al. 1998 ) n Each mode shows specific cell-cycle behavior n n ICA modes remain inactive within some of the experiments Dimension reduction improve a prediction of cellcycle regulated genes

ICA applied to Time courses(2) by Liebermeister

ICA applied to Time courses(3) by Liebermeister

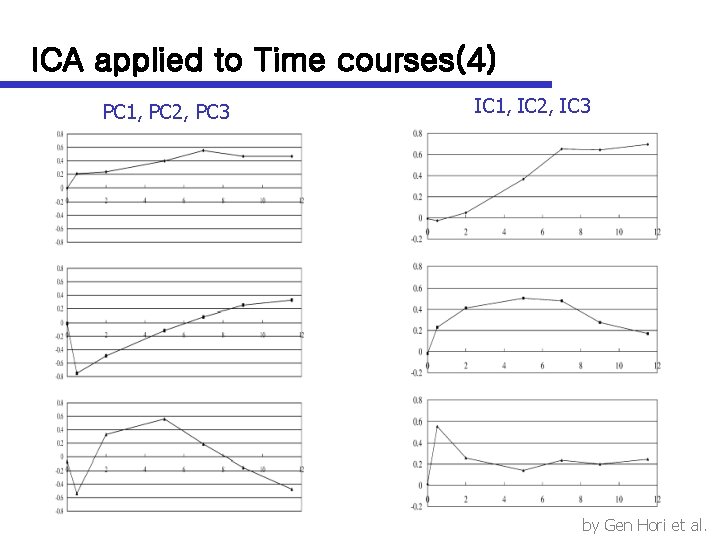
ICA applied to Time courses(4) PC 1, PC 2, PC 3 IC 1, IC 2, IC 3 by Gen Hori et al.

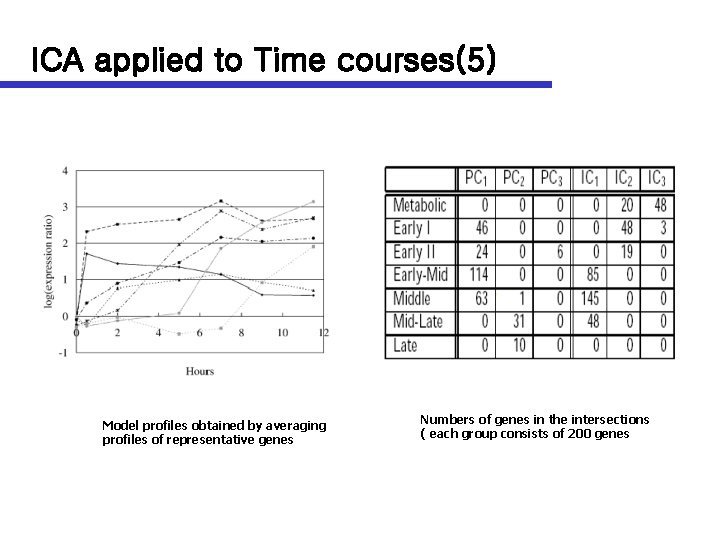
ICA applied to Time courses(5) Model profiles obtained by averaging profiles of representative genes Numbers of genes in the intersections ( each group consists of 200 genes

ICA applied to Different cell types n B-cell lymphoma data ( Alizadeh et al, 2000) n 96 sample 4026 genes n n Samples such as T-cell, activated blood B-cell, leukemia cell lines, etc. Compare the modes to the gene clusters that had been determined in the original work using hierarchical clustering n n Mode 2 and 5 show the highest variances, point to the proliferation and lymphnode gene clusters Mode 8 and 12 are related to ‘pan B-cell’ and the ‘germinal center B-cell’ cluster respectively