Metabolism of amino acids porphyrins PROTEIN TURNOVER Protein
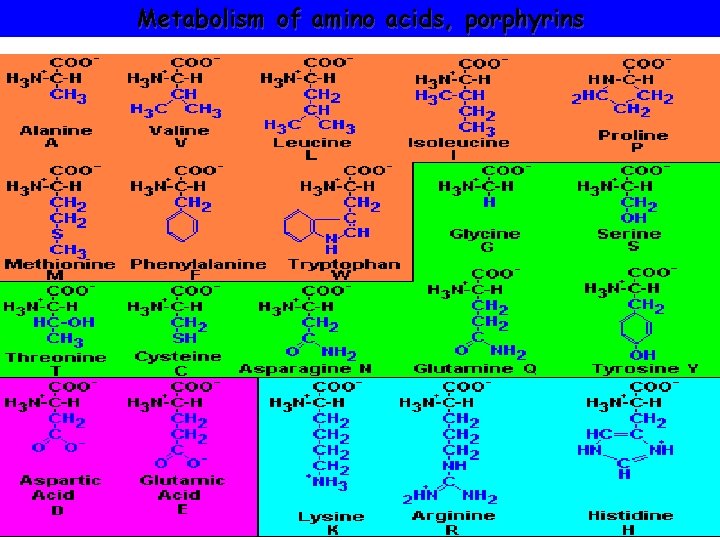
Metabolism of amino acids, porphyrins
PROTEIN TURNOVER Protein turnover — the degradation and resynthesis of proteins Half-lives of proteins – from several minutes to many years Structural proteins – usually stable (lens protein crystallin lives during the whole life of the organism) Regulatory proteins - short lived (altering the amounts of these proteins can rapidly change the rate of metabolic processes) How can a cell distinguish proteins that are meant for degradation?
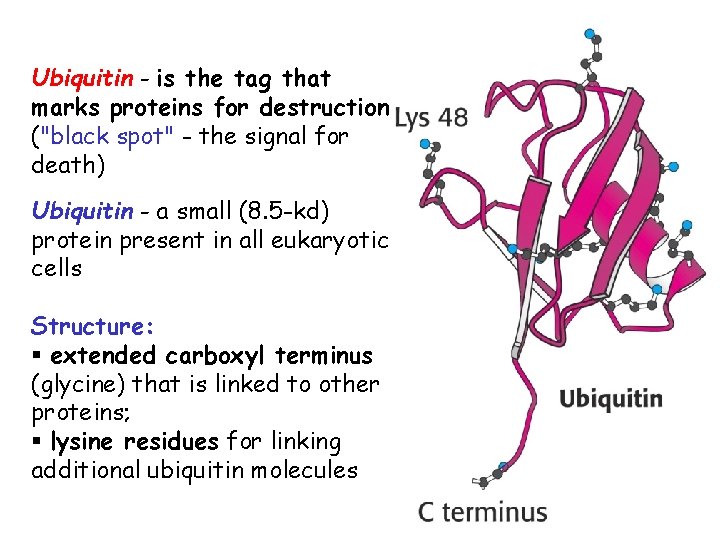
Ubiquitin - is the tag that marks proteins for destruction ("black spot" - the signal for death) Ubiquitin - a small (8. 5 -kd) protein present in all eukaryotic cells Structure: § extended carboxyl terminus (glycine) that is linked to other proteins; § lysine residues for linking additional ubiquitin molecules
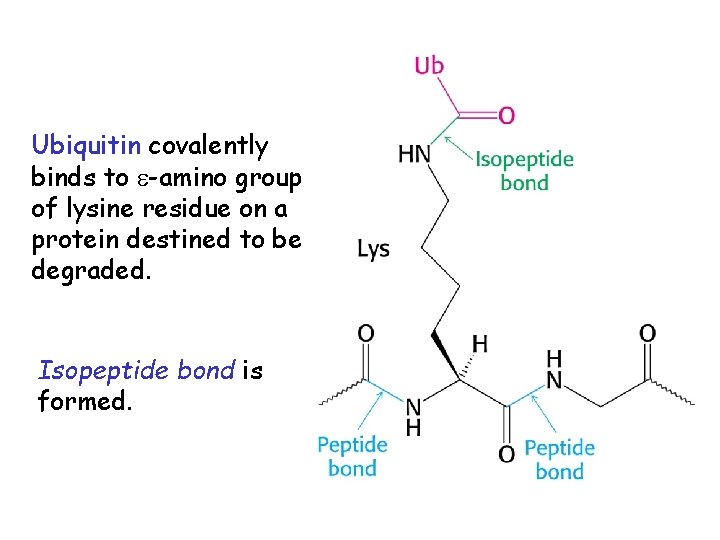
Ubiquitin covalently binds to -amino group of lysine residue on a protein destined to be degraded. Isopeptide bond is formed.
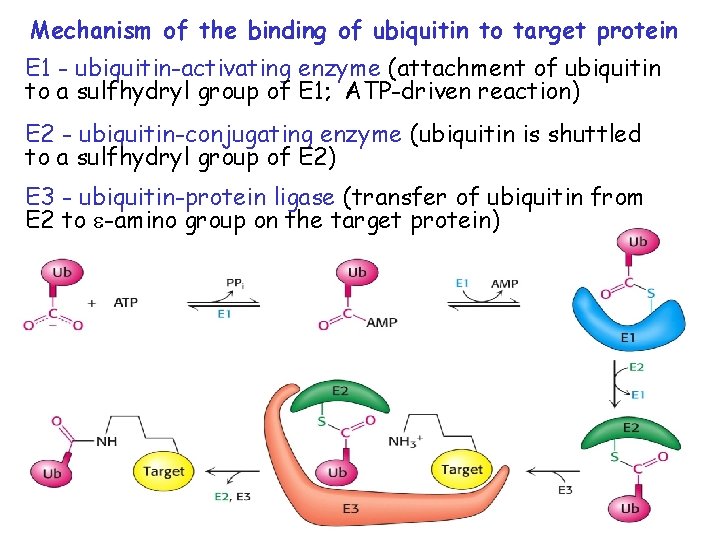
Mechanism of the binding of ubiquitin to target protein E 1 - ubiquitin-activating enzyme (attachment of ubiquitin to a sulfhydryl group of E 1; ATP-driven reaction) E 2 - ubiquitin-conjugating enzyme (ubiquitin is shuttled to a sulfhydryl group of E 2) E 3 - ubiquitin-protein ligase (transfer of ubiquitin from E 2 to -amino group on the target protein)
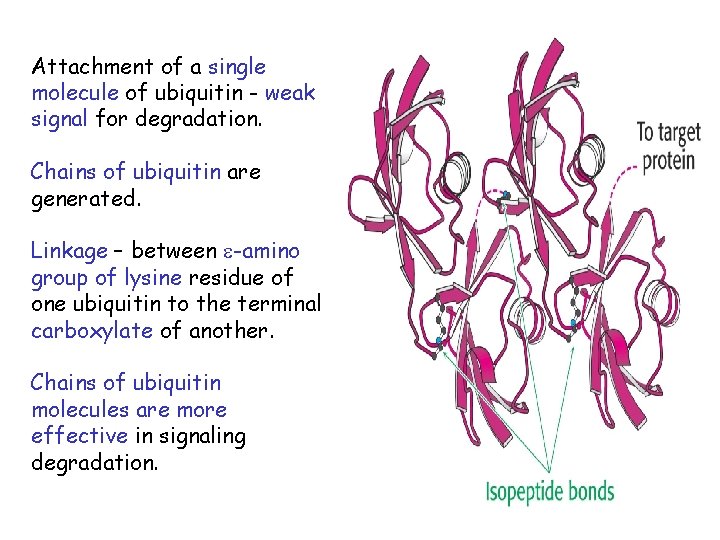
Attachment of a single molecule of ubiquitin - weak signal for degradation. Chains of ubiquitin are generated. Linkage – between -amino group of lysine residue of one ubiquitin to the terminal carboxylate of another. Chains of ubiquitin molecules are more effective in signaling degradation.
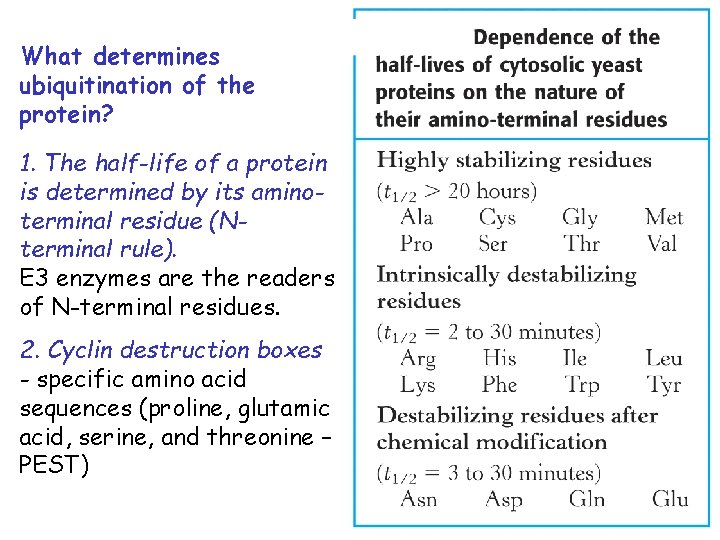
What determines ubiquitination of the protein? 1. The half-life of a protein is determined by its aminoterminal residue (Nterminal rule). E 3 enzymes are the readers of N-terminal residues. 2. Cyclin destruction boxes - specific amino acid sequences (proline, glutamic acid, serine, and threonine – PEST)
Digestion of the Ubiquitin-Tagged Proteins What is the executioner of the protein death? A large protease complex proteasome or the 26 S proteasome digests the ubiquitinated proteins. 26 S proteasome - ATP-driven multisubunit protease. 26 S proteasome consists of two components: § 20 S - catalytic subunit § 19 S - regulatory subunit
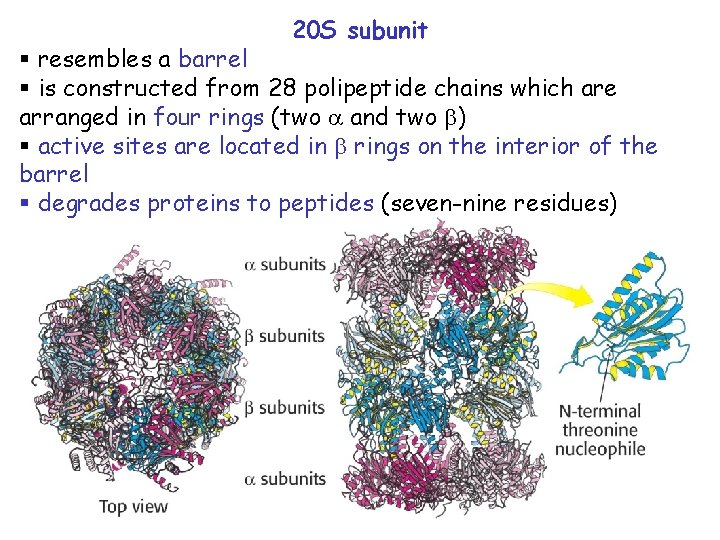
20 S subunit § resembles a barrel § is constructed from 28 polipeptide chains which are arranged in four rings (two and two ) § active sites are located in rings on the interior of the barrel § degrades proteins to peptides (seven-nine residues)
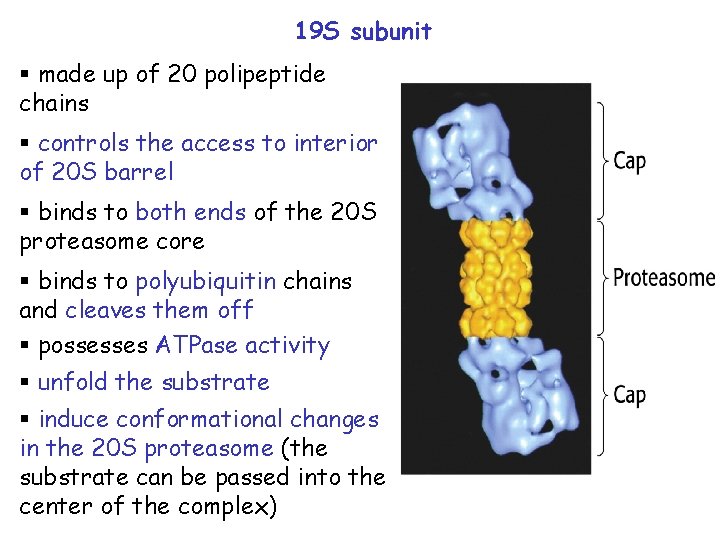
19 S subunit § made up of 20 polipeptide chains § controls the access to interior of 20 S barrel § binds to both ends of the 20 S proteasome core § binds to polyubiquitin chains and cleaves them off § possesses ATPase activity § unfold the substrate § induce conformational changes in the 20 S proteasome (the substrate can be passed into the center of the complex)
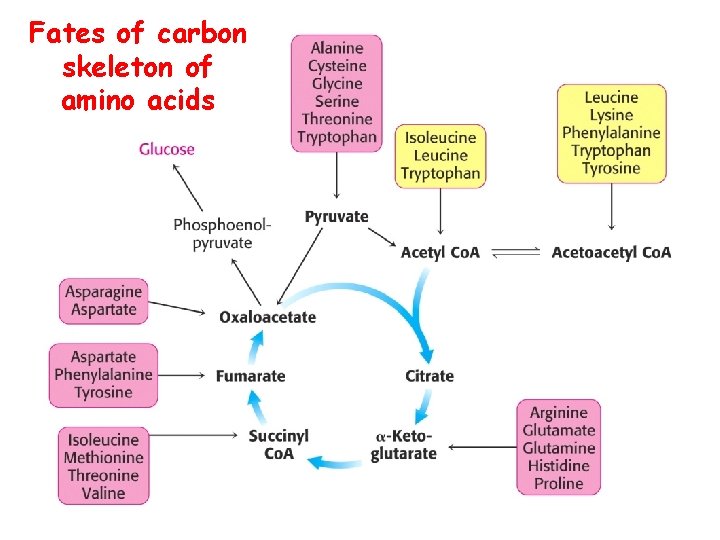
Fates of carbon skeleton of amino acids
Glucogenic vs ketogenic amino acids • Glucogenic amino acids (are degraded to pyruvate or citric acid cycle intermediates) can supply gluconeogenesis pathway • Ketogenic amino acids (are degraded to acetyl Co. A or acetoacetyl Co. A) - can contribute to synthesis of fatty acids or ketone bodies • Some amino acids are both glucogenic and ketogenic
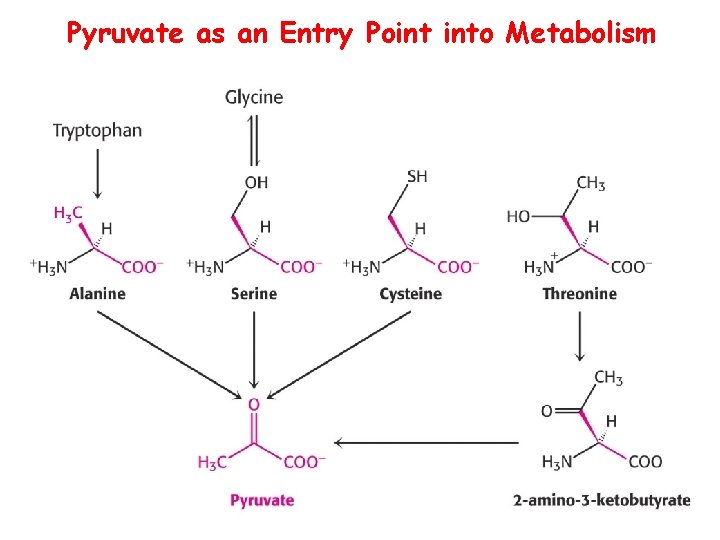
Pyruvate as an Entry Point into Metabolism
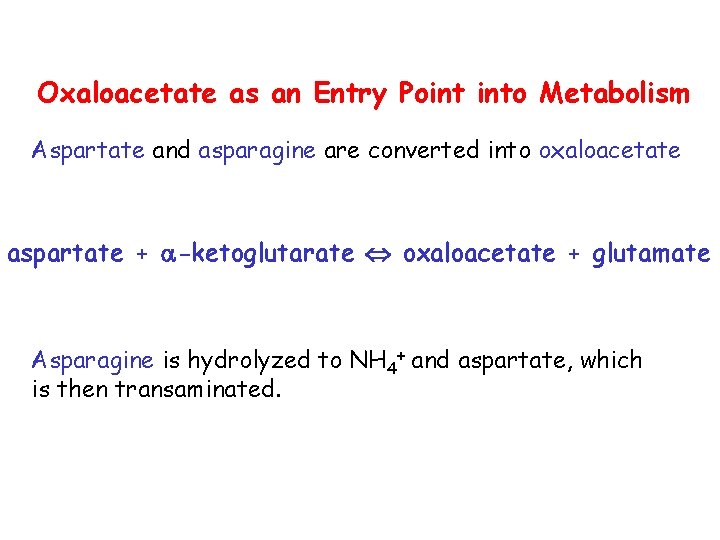
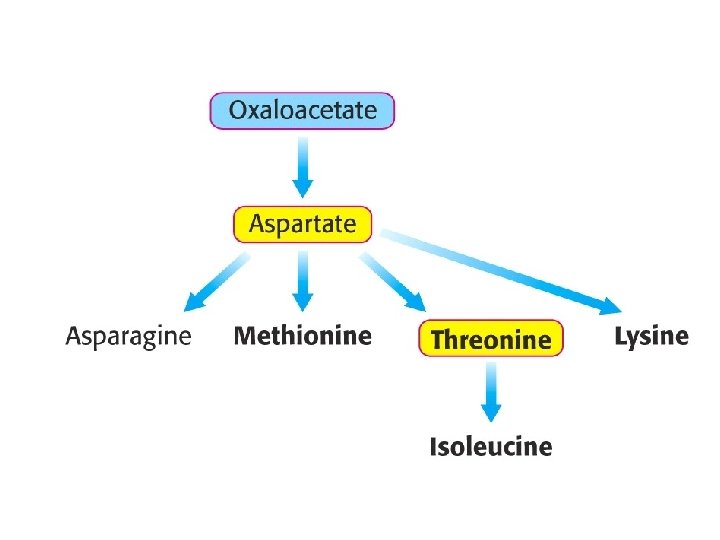
Oxaloacetate as an Entry Point into Metabolism Aspartate and asparagine are converted into oxaloacetate aspartate + -ketoglutarate oxaloacetate + glutamate Asparagine is hydrolyzed to NH 4+ and aspartate, which is then transaminated.
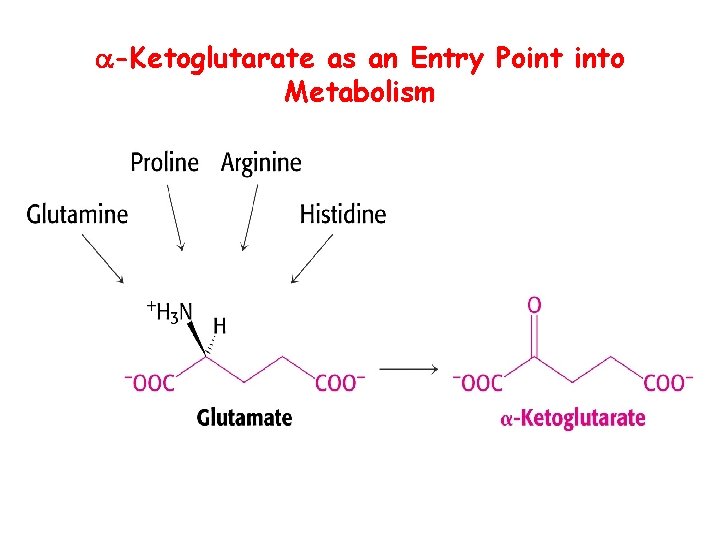
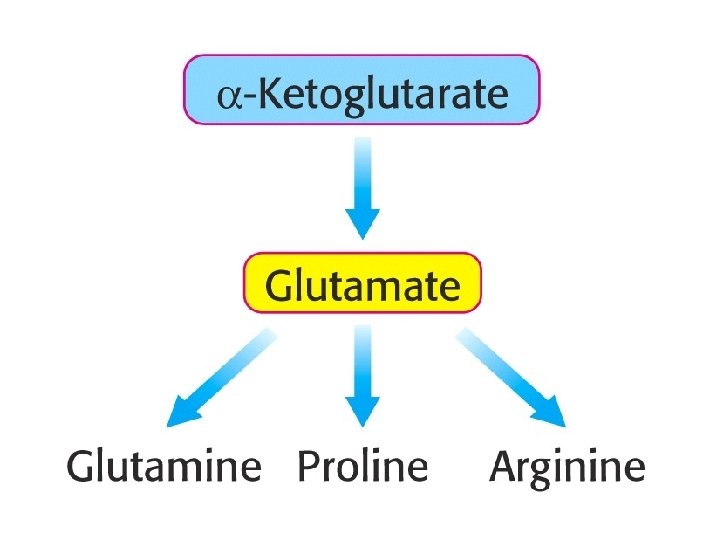
-Ketoglutarate as an Entry Point into Metabolism
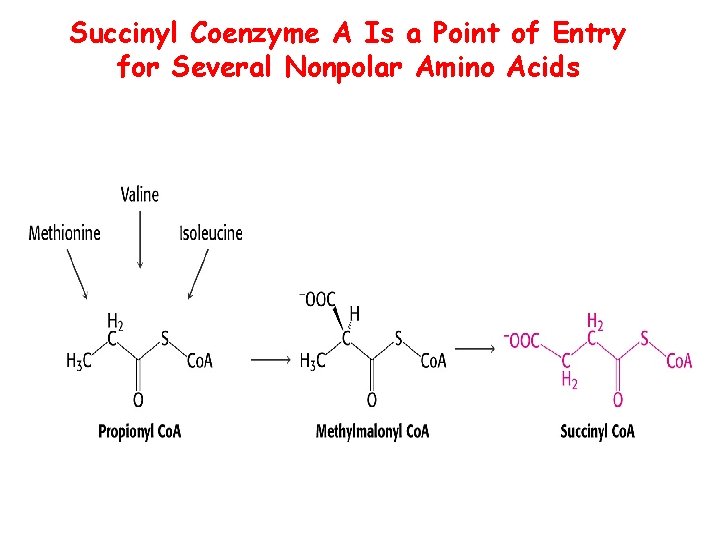
Succinyl Coenzyme A Is a Point of Entry for Several Nonpolar Amino Acids
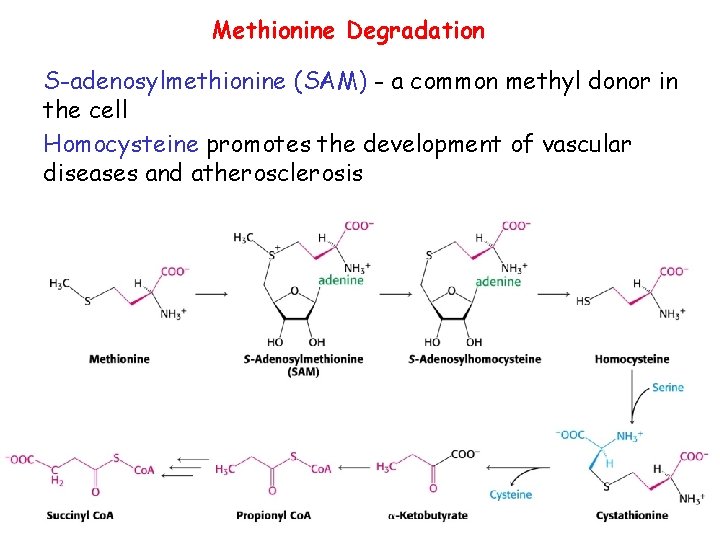
Methionine Degradation S-adenosylmethionine (SAM) - a common methyl donor in the cell Homocysteine promotes the development of vascular diseases and atherosclerosis
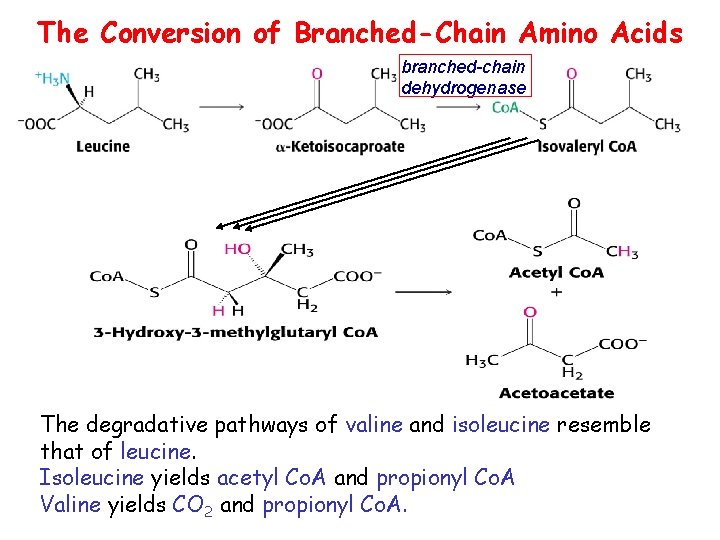
The Conversion of Branched-Chain Amino Acids branched-chain dehydrogenase The degradative pathways of valine and isoleucine resemble that of leucine. Isoleucine yields acetyl Co. A and propionyl Co. A Valine yields CO 2 and propionyl Co. A.
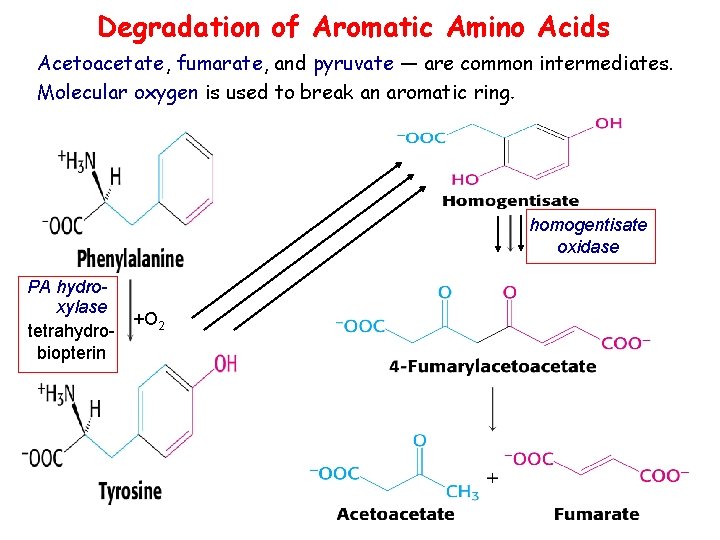
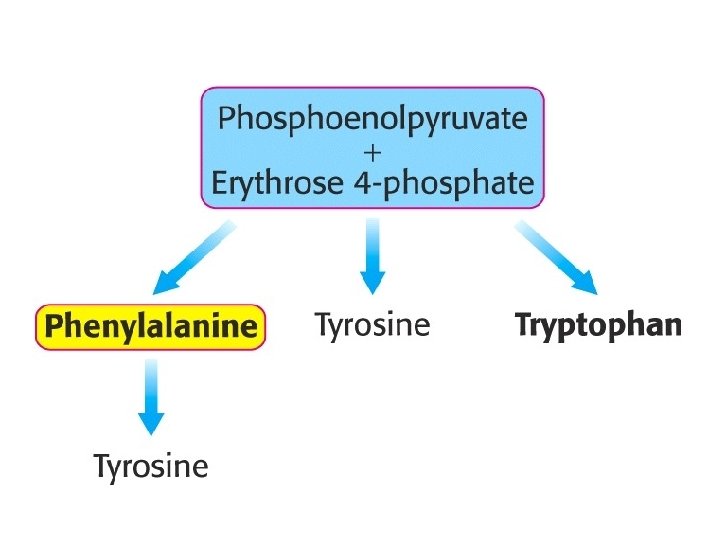
Degradation of Aromatic Amino Acids Acetoacetate, fumarate, and pyruvate — are common intermediates. Molecular oxygen is used to break an aromatic ring. homogentisate oxidase PA hydroxylase +O 2 tetrahydrobiopterin
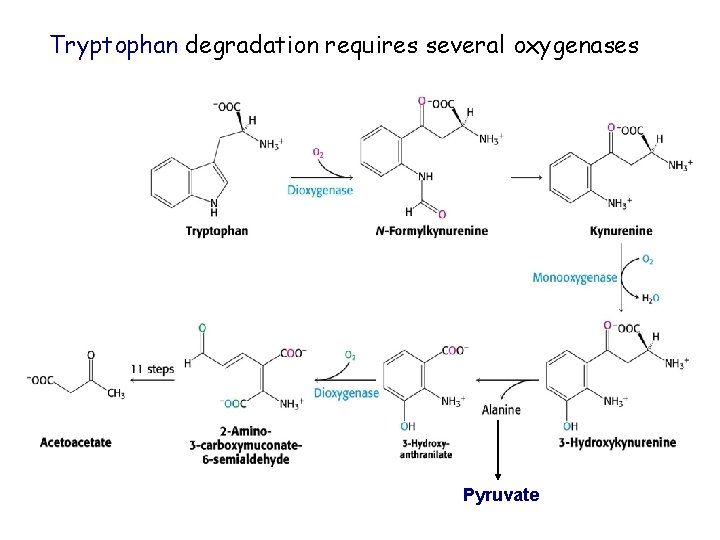
Tryptophan degradation requires several oxygenases Pyruvate

SYNTHESIS OF NITRIC OXIDE (NO) FROM ARGININE • Nitric oxide (. N=O) is a gas which can diffuse rapidly into cells, and is a messenger that activates guanylyl cyclase (GMP synthesis) • NO relaxes blood vessels, lowers blood pressure, and is a neurotransmitter in the brain • Nitroglycerin is converted to NO and dilates coronary arteries in treating angina pectoris
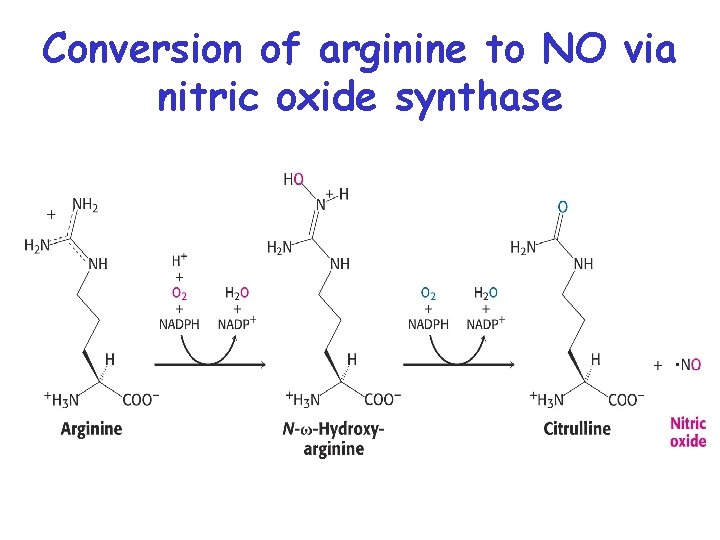
Conversion of arginine to NO via nitric oxide synthase

• Plants and microorganisms can make all 20 amino acids • Humans can make only 11 of the 20 amino acids (“nonessential” amino acids) • Nonessential amino acids for mammals are usually derived from intermediates of glycolysis or the citric acid cycle • The others are classed as "essential" amino acids and must be obtained in the diet
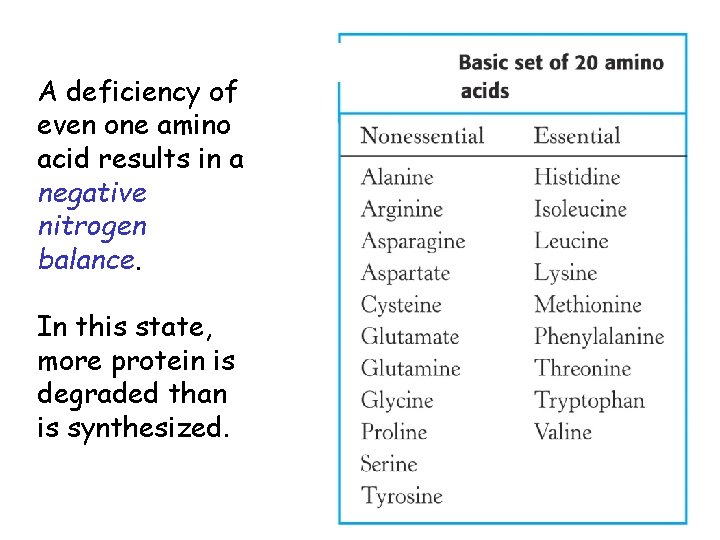
A deficiency of even one amino acid results in a negative nitrogen balance. In this state, more protein is degraded than is synthesized.
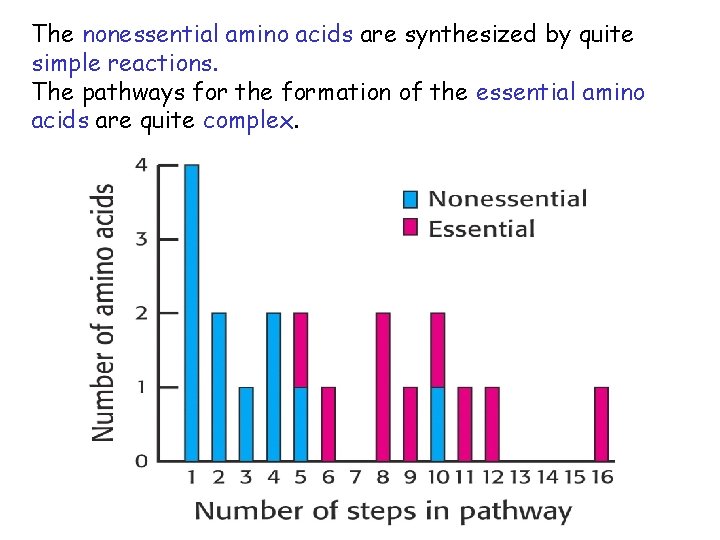
The nonessential amino acids are synthesized by quite simple reactions. The pathways for the formation of the essential amino acids are quite complex.
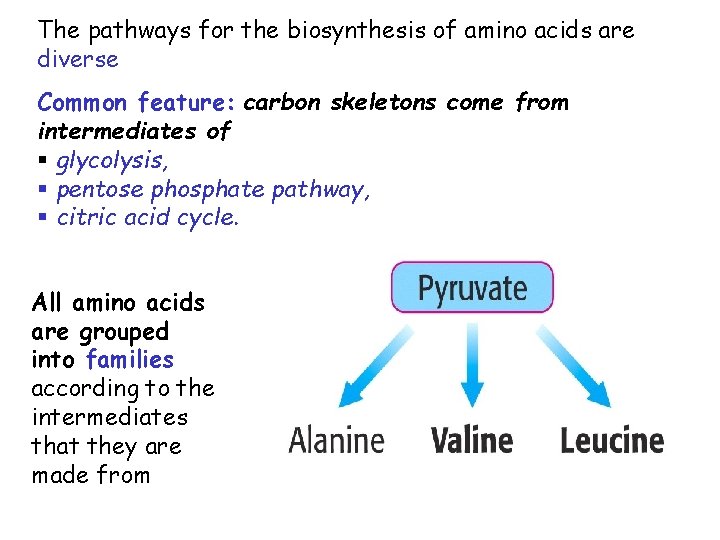
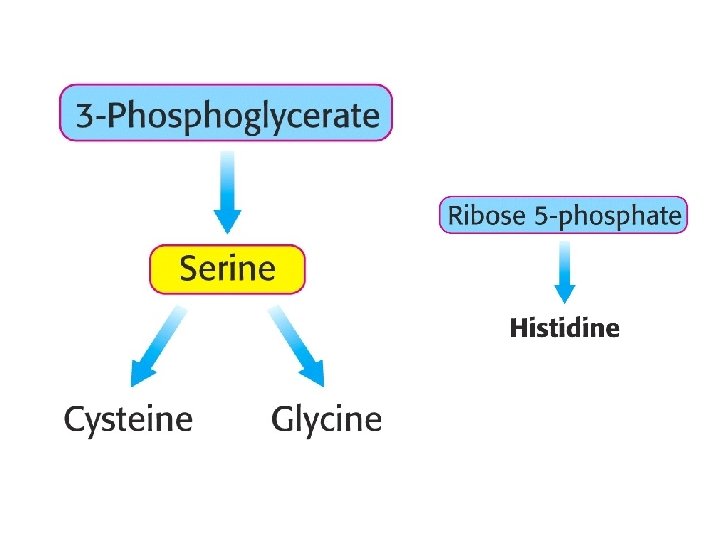
The pathways for the biosynthesis of amino acids are diverse Common feature: carbon skeletons come from intermediates of § glycolysis, § pentose phosphate pathway, § citric acid cycle. All amino acids are grouped into families according to the intermediates that they are made from
- Slides: 30