Membranes Structure of Membrane Proteins The Manifold Roles

Membranes Structure of Membrane Proteins
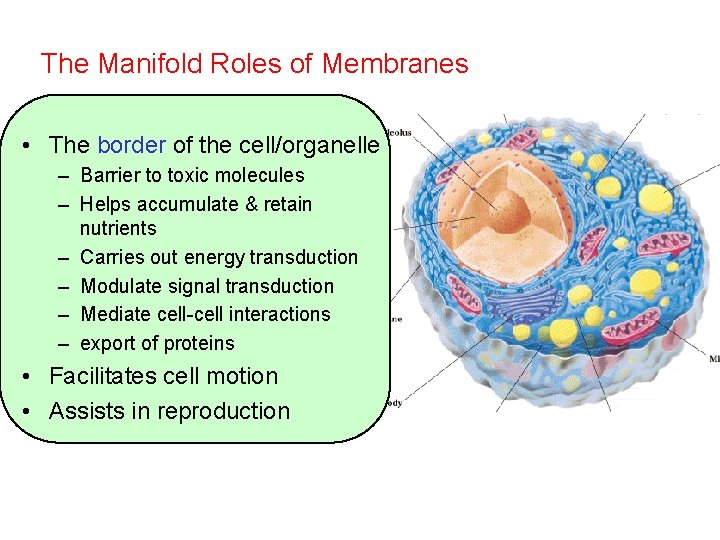
The Manifold Roles of Membranes • The border of the cell/organelle – Barrier to toxic molecules – Helps accumulate & retain nutrients – Carries out energy transduction – Modulate signal transduction – Mediate cell-cell interactions – export of proteins • Facilitates cell motion • Assists in reproduction
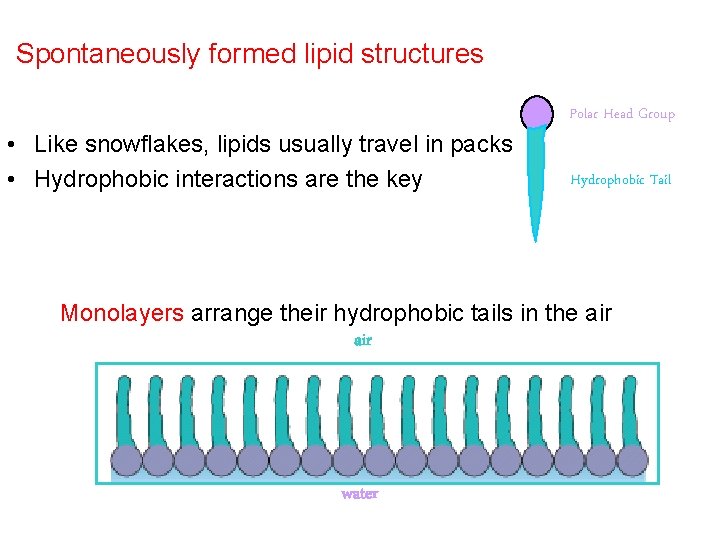
Spontaneously formed lipid structures • Like snowflakes, lipids usually travel in packs • Hydrophobic interactions are the key Polar Head Group Hydrophobic Tail Monolayers arrange their hydrophobic tails in the air water
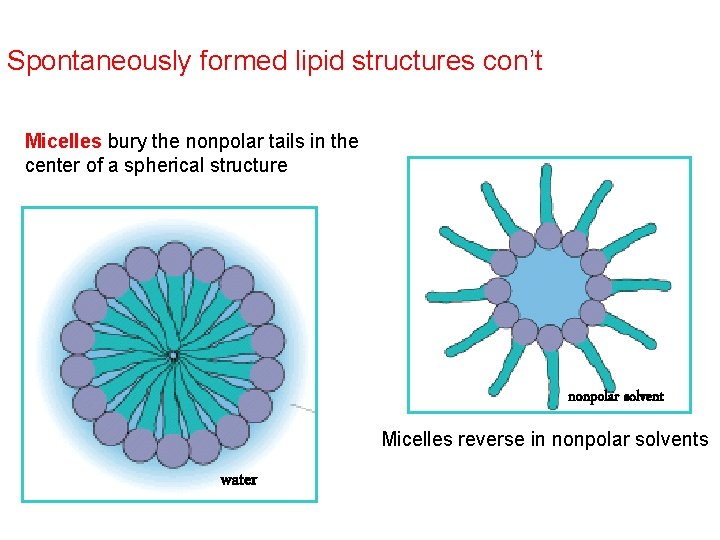
Spontaneously formed lipid structures con’t Micelles bury the nonpolar tails in the center of a spherical structure nonpolar solvent Micelles reverse in nonpolar solvents water
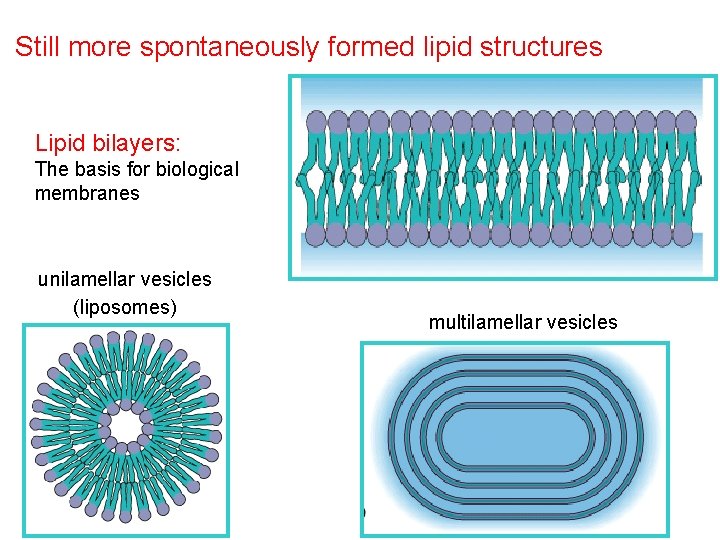
Still more spontaneously formed lipid structures Lipid bilayers: The basis for biological membranes unilamellar vesicles (liposomes) multilamellar vesicles

The Fluid Mosaic Model of Singer & Nicholson • • The phospholipid bilayer is a fluid matrix The bilayer is a two-dimensional solvent Lipids and proteins can undergo rotational and lateral movement Two classes of proteins: – peripheral proteins (extrinsic proteins) – integral proteins (intrinsic proteins)
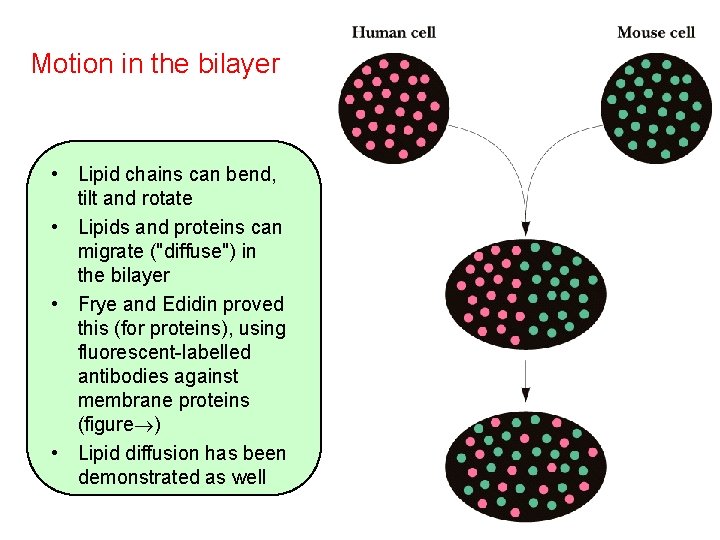
Motion in the bilayer • Lipid chains can bend, tilt and rotate • Lipids and proteins can migrate ("diffuse") in the bilayer • Frye and Edidin proved this (for proteins), using fluorescent-labelled antibodies against membrane proteins (figure ) • Lipid diffusion has been demonstrated as well
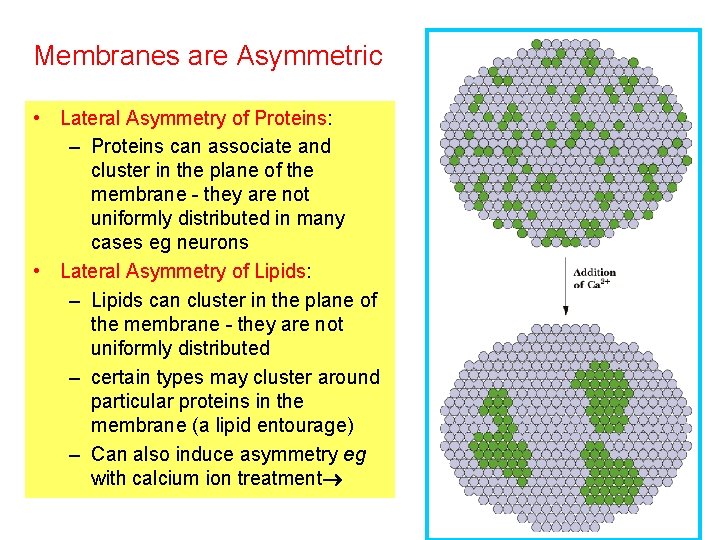
Membranes are Asymmetric • Lateral Asymmetry of Proteins: – Proteins can associate and cluster in the plane of the membrane - they are not uniformly distributed in many cases eg neurons • Lateral Asymmetry of Lipids: – Lipids can cluster in the plane of the membrane - they are not uniformly distributed – certain types may cluster around particular proteins in the membrane (a lipid entourage) – Can also induce asymmetry eg with calcium ion treatment

Transverse Asymmetry of Membranes • Functions of membrane proteins depends on their orientation within the membrane • Membrane proteins are not tossed into the membrane randomly but have a specific topology eg Glycophorin Outside Inside

Transverse asymmetry of membrane lipids In most cell membranes, the lipid composition of the outer monolayer differs from that of the inner monolayer: phosphatidylcholine phosphatidyethanolamine phosphatidylserine sphingomyelin total percentage of phospholipid
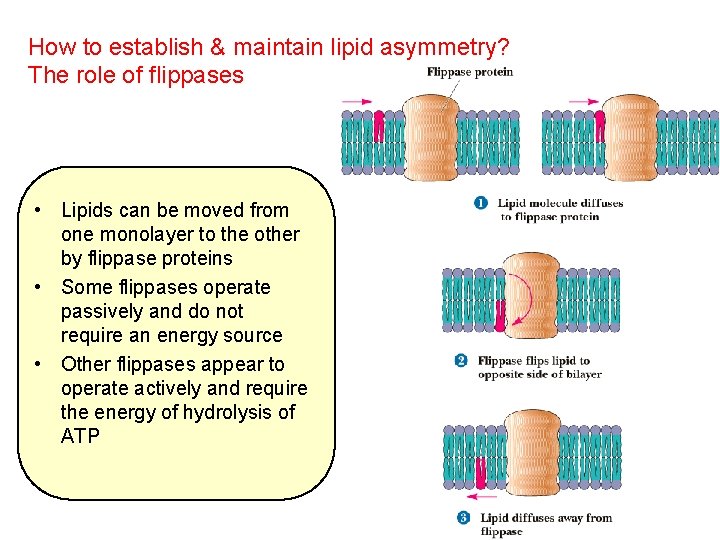
How to establish & maintain lipid asymmetry? The role of flippases • Lipids can be moved from one monolayer to the other by flippase proteins • Some flippases operate passively and do not require an energy source • Other flippases appear to operate actively and require the energy of hydrolysis of ATP

Membrane Phase Transitions: The "melting" of membrane lipids • Below a certain transition temperature, membrane lipids are rigid & tightly packed (Gel Phase) • Above the transition temperature, lipids are more flexible and mobile (Liquid crystal phase) • The transition temperature is characteristic of the lipids in the membrane • Only pure lipid systems give sharp, well-defined transition temperatures Gel Liquid crystal

Observing Membrane Phase Transitions by Calorimetry Transition Temperature Liquid crystal Heat absorbed Gel Anti conformation Gauche conformations Temperature
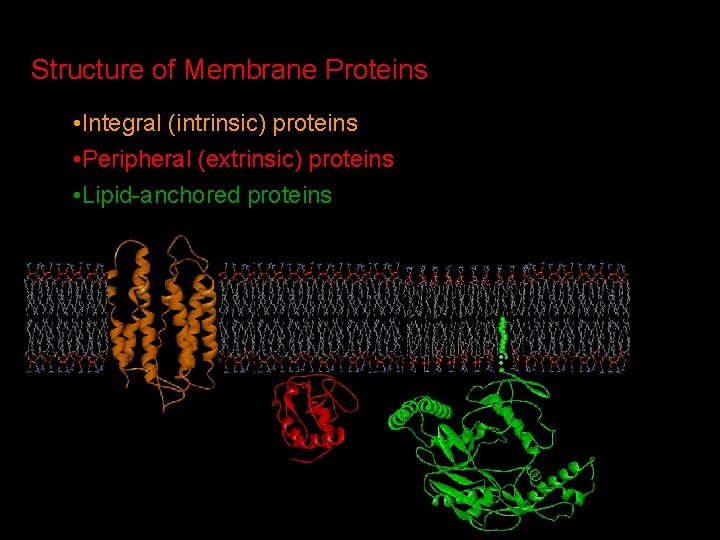
Structure of Membrane Proteins • Integral (intrinsic) proteins • Peripheral (extrinsic) proteins • Lipid-anchored proteins

Peripheral Membrane Proteins • Not strongly bound to the membrane • Can be dissociated with mild detergent treatment or with high salt concentrations • What holds them there in the first place?

Integral Membrane Proteins (IMPs) • Strongly imbedded in the bilayer • Can only be removed by disrupting the membrane (eg detergents) • Often span the membrane (transmembrane) • “Inside out” compared to other globular proteins. . meaning?
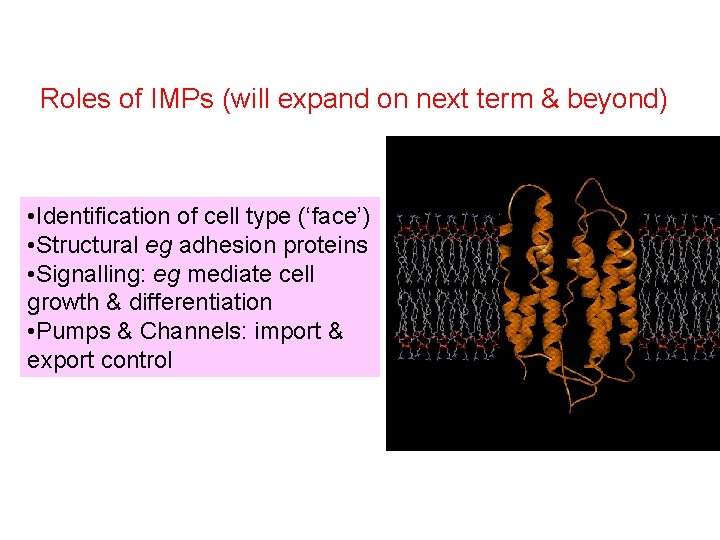
Roles of IMPs (will expand on next term & beyond) • Identification of cell type (‘face’) • Structural eg adhesion proteins • Signalling: eg mediate cell growth & differentiation • Pumps & Channels: import & export control
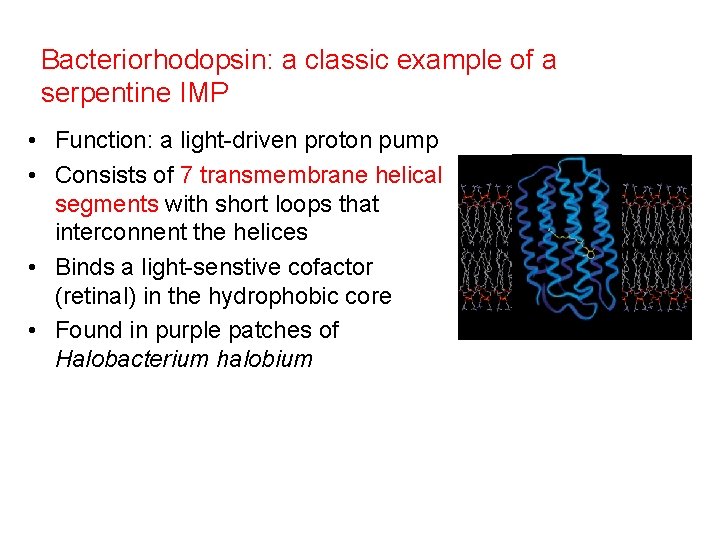
Bacteriorhodopsin: a classic example of a serpentine IMP • Function: a light-driven proton pump • Consists of 7 transmembrane helical segments with short loops that interconnent the helices • Binds a light-senstive cofactor (retinal) in the hydrophobic core • Found in purple patches of Halobacterium halobium
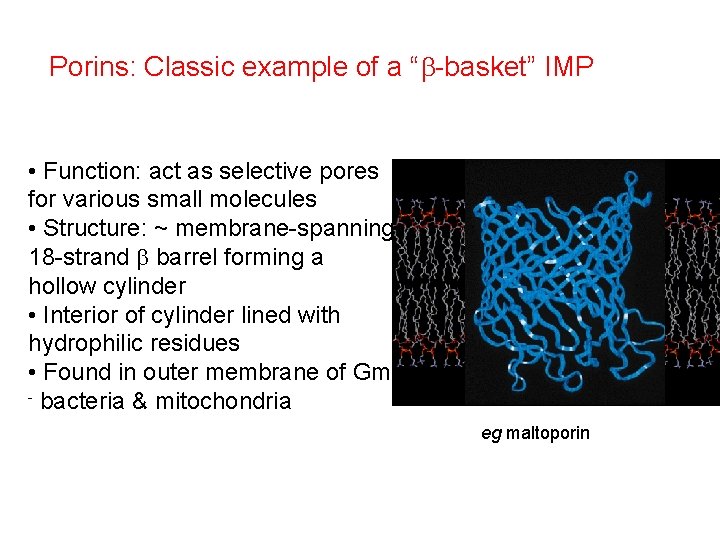
Porins: Classic example of a “b-basket” IMP • Function: act as selective pores for various small molecules • Structure: ~ membrane-spanning 18 -strand b barrel forming a hollow cylinder • Interior of cylinder lined with hydrophilic residues • Found in outer membrane of Gm - bacteria & mitochondria eg maltoporin
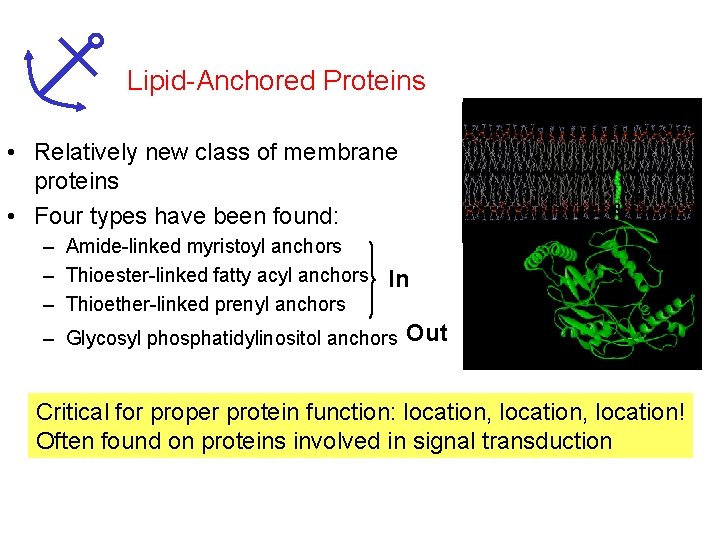
Lipid-Anchored Proteins • Relatively new class of membrane proteins • Four types have been found: – Amide-linked myristoyl anchors – Thioester-linked fatty acyl anchors In – Thioether-linked prenyl anchors – Glycosyl phosphatidylinositol anchors Out Critical for proper protein function: location, location! Often found on proteins involved in signal transduction
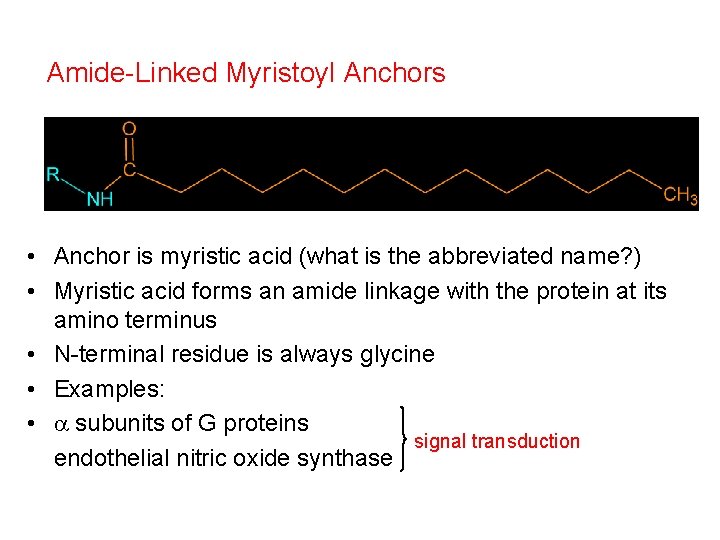
Amide-Linked Myristoyl Anchors • Anchor is myristic acid (what is the abbreviated name? ) • Myristic acid forms an amide linkage with the protein at its amino terminus • N-terminal residue is always glycine • Examples: • a subunits of G proteins signal transduction endothelial nitric oxide synthase
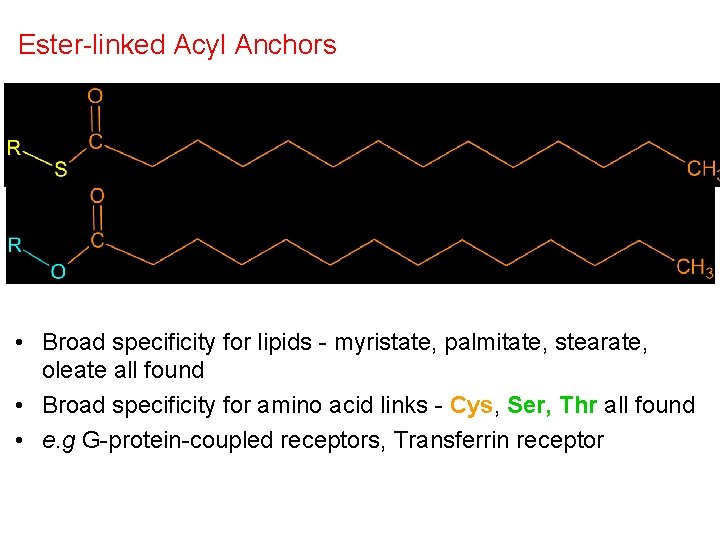
Ester-linked Acyl Anchors • Broad specificity for lipids - myristate, palmitate, stearate, oleate all found • Broad specificity for amino acid links - Cys, Ser, Thr all found • e. g G-protein-coupled receptors, Transferrin receptor
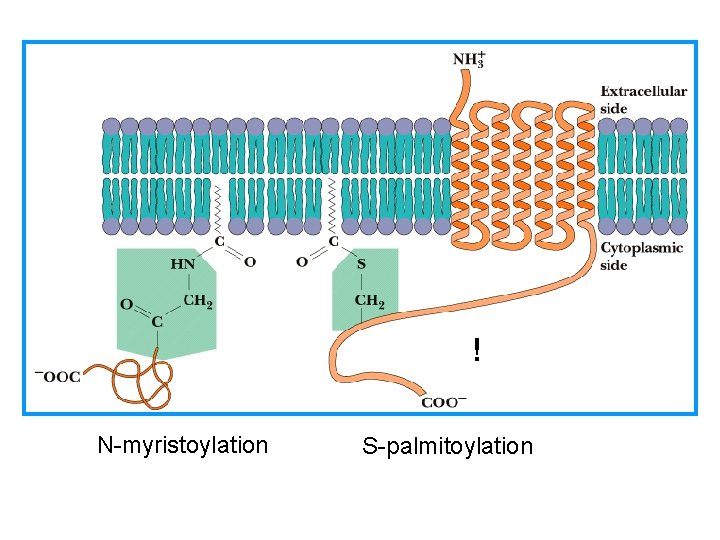
! N-myristoylation S-palmitoylation
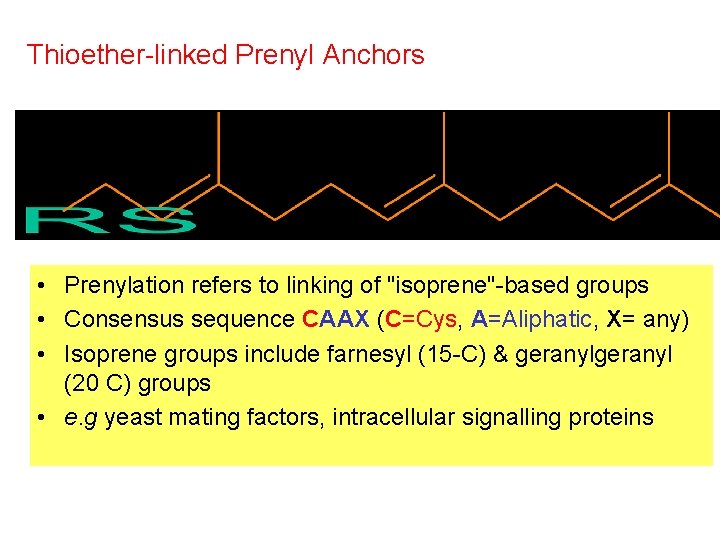
Thioether-linked Prenyl Anchors • Prenylation refers to linking of "isoprene"-based groups • Consensus sequence CAAX (C=Cys, A=Aliphatic, X= any) • Isoprene groups include farnesyl (15 -C) & geranyl (20 C) groups • e. g yeast mating factors, intracellular signalling proteins
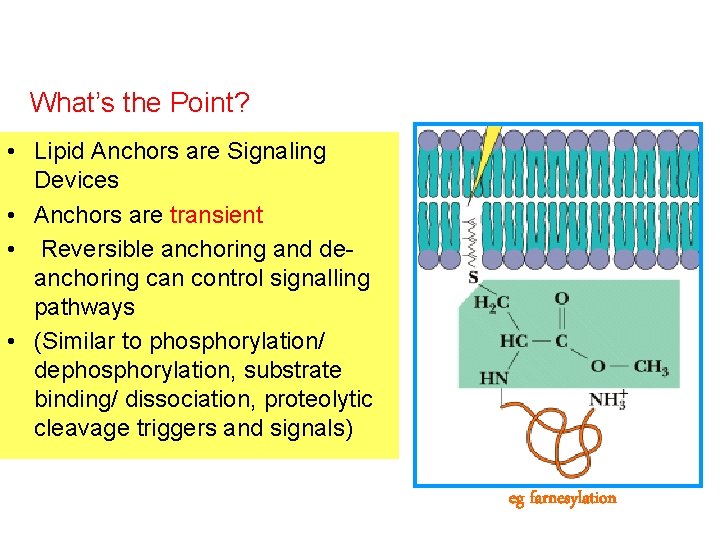
What’s the Point? • Lipid Anchors are Signaling Devices • Anchors are transient • Reversible anchoring and deanchoring can control signalling pathways • (Similar to phosphorylation/ dephosphorylation, substrate binding/ dissociation, proteolytic cleavage triggers and signals) eg farnesylation
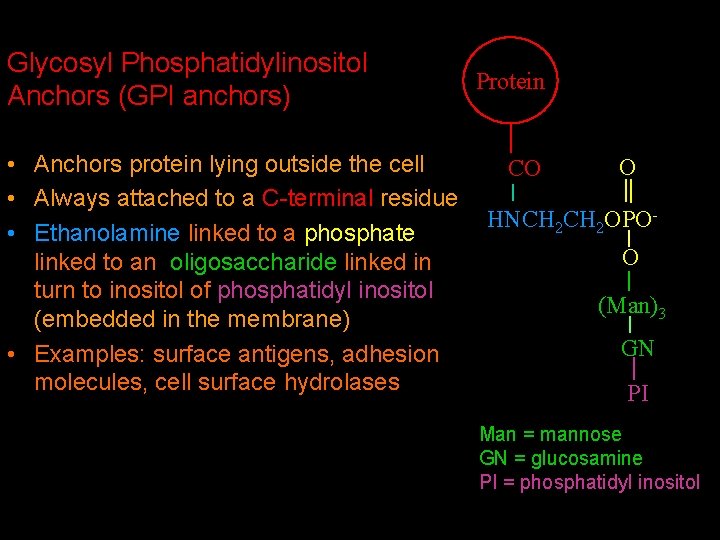
Glycosyl Phosphatidylinositol Anchors (GPI anchors) • Anchors protein lying outside the cell • Always attached to a C-terminal residue • Ethanolamine linked to a phosphate linked to an oligosaccharide linked in turn to inositol of phosphatidyl inositol (embedded in the membrane) • Examples: surface antigens, adhesion molecules, cell surface hydrolases Protein CO C O HNCH 2 OPOO (Man)3 GN PI Man = mannose GN = glucosamine PI = phosphatidyl inositol
- Slides: 26