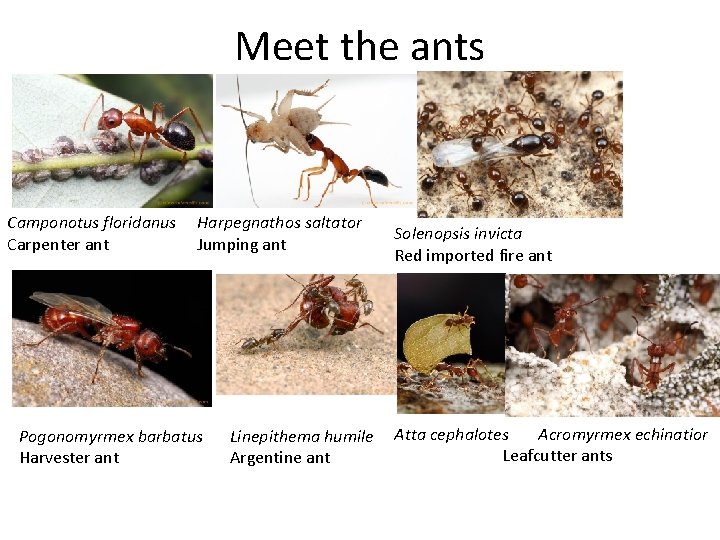
Meet the ants Camponotus floridanus Carpenter ant Harpegnathos




- Slides: 11

Meet the ants Camponotus floridanus Carpenter ant Harpegnathos saltator Jumping ant Pogonomyrmex barbatus Harvester ant Linepithema humile Argentine ant Solenopsis invicta Red imported fire ant Atta cephalotes Acromyrmex echinatior Leafcutter ants

Now meet their genomes…

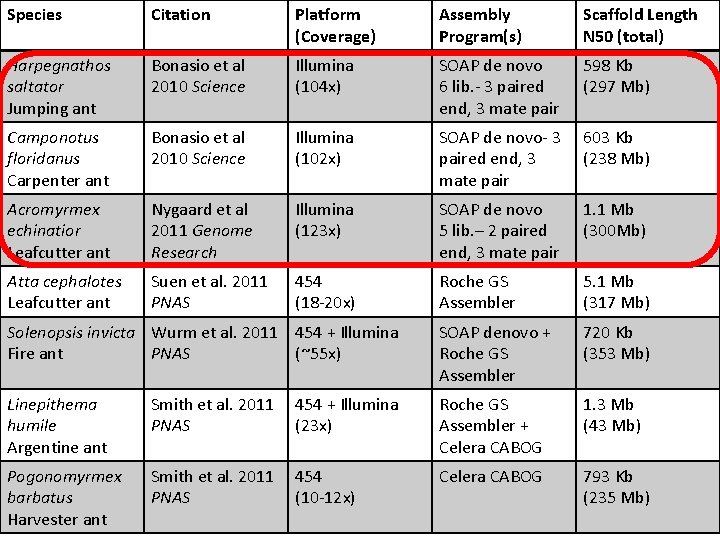
Species Citation Platform (Coverage) Assembly Program(s) Scaffold Length N 50 (total) Harpegnathos saltator Jumping ant Bonasio et al 2010 Science Illumina (104 x) SOAP de novo 6 lib. - 3 paired end, 3 mate pair 598 Kb (297 Mb) Camponotus floridanus Carpenter ant Bonasio et al 2010 Science Illumina (102 x) SOAP de novo- 3 paired end, 3 mate pair 603 Kb (238 Mb) Acromyrmex echinatior Leafcutter ant Nygaard et al 2011 Genome Research Illumina (123 x) SOAP de novo 5 lib. – 2 paired end, 3 mate pair 1. 1 Mb (300 Mb) Atta cephalotes Leafcutter ant Suen et al. 2011 PNAS 454 (18 -20 x) Roche GS Assembler 5. 1 Mb (317 Mb) Solenopsis invicta Wurm et al. 2011 454 + Illumina Fire ant PNAS (~55 x) SOAP denovo + Roche GS Assembler 720 Kb (353 Mb) Linepithema humile Argentine ant Smith et al. 2011 PNAS 454 + Illumina (23 x) Roche GS Assembler + Celera CABOG 1. 3 Mb (43 Mb) Pogonomyrmex barbatus Harvester ant Smith et al. 2011 PNAS 454 (10 -12 x) Celera CABOG 793 Kb (235 Mb)

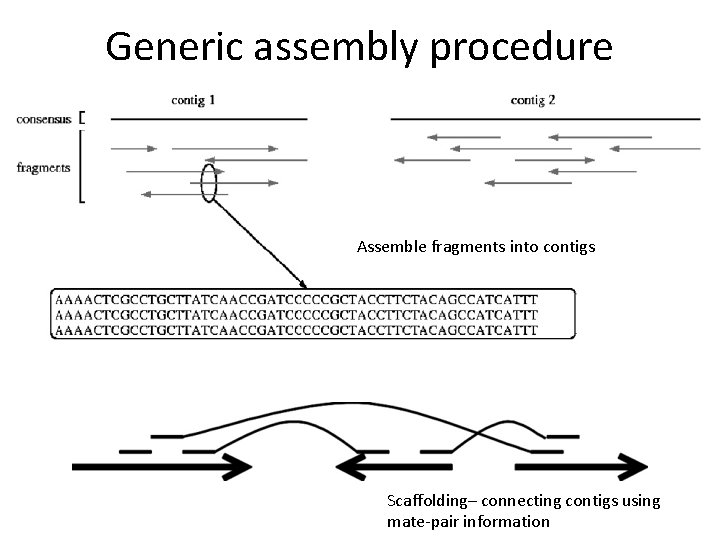
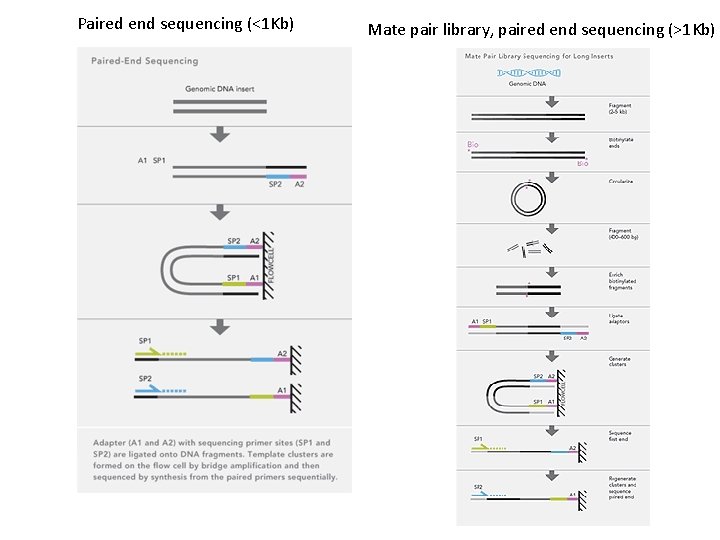
Generic assembly procedure Assemble fragments into contigs Scaffolding– connecting contigs using mate-pair information

Steps involved in Illumina Assembly 1) Download data (qseq file– sequences with quality scores) 2) Filter data A) Filter low quality reads B) Trim adapter sequences 3) SOAPdenovo steps A) Preassembly error correction (Identify pairs of reads sharing a common sequence (k-mer, e. g. 17 -20), estimate k-mer frequency, and remove erroneous k-mers) B) Construct contigs based on short insert libraries (200 -800 bp) C) Join contigs into scaffolds using information from large insert mate pair libraries (1 Kb-10 Kb) D) Do local reassembly of unresolved gap regions using Gap Closer for SOAPdenovo

2) Filtering data (specifics) • A) Remove low quality reads – Remove reads that do not pass GA analysis Failed_Chastity filter (have an N in the last column of the GA export file) – Can use R Bio. Conductor Short. Read package (may have to convert files from qseq to fastq format) • B) Remove adapter sequences – need adapter sequence information from person that did sequencing – Can use vectorstrip in EMBOSS

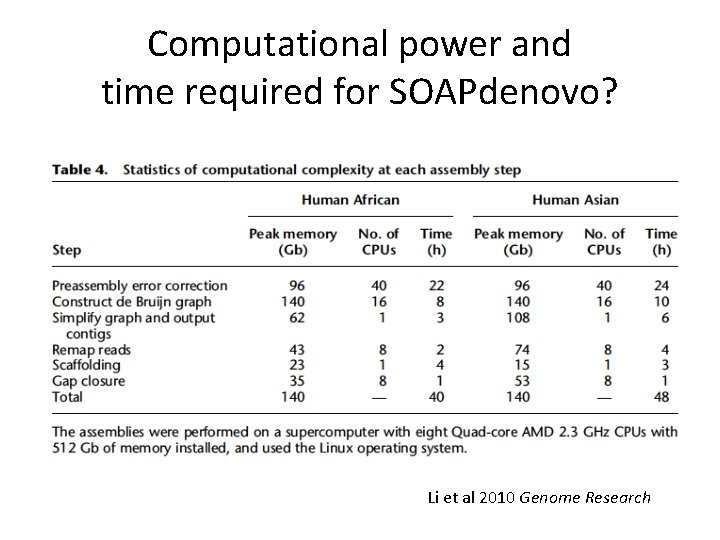
Computational power and time required for SOAPdenovo? Li et al 2010 Genome Research

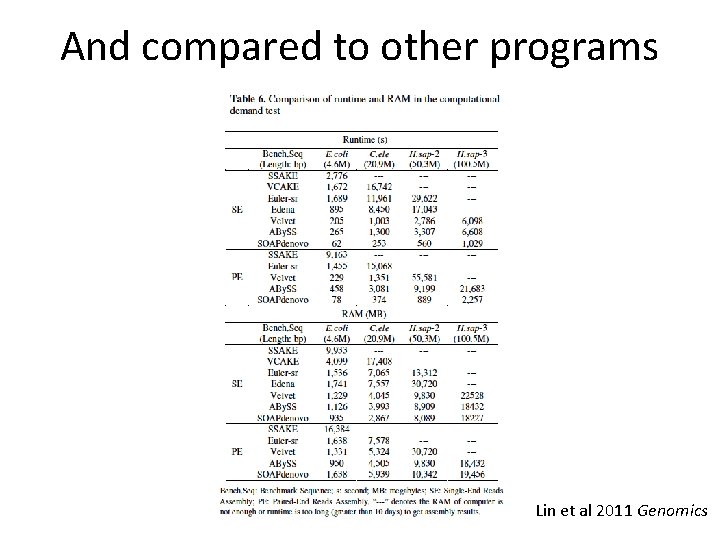
And compared to other programs Lin et al 2011 Genomics

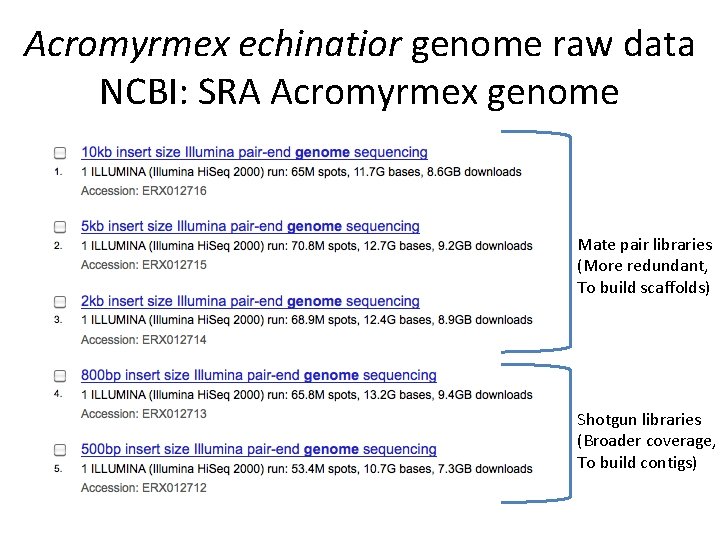
Acromyrmex echinatior genome raw data NCBI: SRA Acromyrmex genome Mate pair libraries (More redundant, To build scaffolds) Shotgun libraries (Broader coverage, To build contigs)


Paired end sequencing (<1 Kb) Mate pair library, paired end sequencing (>1 Kb)