Medical Image Processing with IDL Medical Image Processing





































- Slides: 87

Medical Image Processing with IDL

Medical Image Processing with IDL This is a three-day course on image processing in IDL, with an emphasis on hands-on demonstration of image processing techniques commonly used in medical research. Central to the course is an explanation of how to use IDL’s DICOM toolkit to read, write, copy and query DICOM format files, through several examples and exercises. Participants should be familiar with the IDL programming language, having completed RSI’s Introduction to IDL course, or having at least 3 months experience using IDL.

Course Objectives 1. IDL and the DICOM standard (Day 1) – Overview of the DICOM standard – Introduction to IDL's DICOM toolkit – Examples of querying, reading, writing and cloning DICOM files with IDL 2. Image processing in IDL (Day 2 -3) – Diagnostic and statistical routines – Pixel operations – Neighborhood operations – Global transforms – Geometric operations – Feature extraction

Part I: IDL and the DICOM Standard

The DICOM Standard DICOM (Digital Imaging and Communications in Medicine) was developed in response to the growing need for a standard format for digital imagery produced by instruments from various vendors. The DICOM standard is intended to – promote communication of digital imagery, regardless of device manufacturer – facilitate development of PACS that can interface with other hospital systems – allow creation of databases that can be interrogated by a wide variety of devices distributed geographically. The DICOM standard is currently in version 3. It is documented in 17 "parts", which can be downloaded from the DICOM homepage: http: //medical. nema. org.

The DICOM Standard Part 1: Introduction and Overview Part 2: Conformance Part 3: Information Object Definitions Part 4: Service Class Specifications Part 5: Data Structure and Semantics Part 6: Data Dictionary Part 7: Message Exchange Part 8: Network Communication Support for Message Exchange Part 9: Point to Point Communication Support for Message Exchange Part 10: Media Storage and File Format for Media Interchange Part 11: Media Storage Application Profiles Part 12: Media Formats and Physical Media for Media Interchange Part 14: Grayscale Standard Display Function Part 15: Security and System Management Profiles Part 16: Content Mapping Resource Part 17: Explanatory Information Part 18: Web Access to DICOM Persistent Objects

The DICOM Standard Diagnostic imaging modalities using DICOM today include – NM - Nuclear Medicine – PET - Positron Emission Tomography – MR - Magnetic Resonance – XR - X-Ray – US - Ultrasound – CT - Computed Tomography – RT - Radiation Therapy – CR - Computed Radiography

DICOM Files DICOM Header patient info, modality, image size, etc. A file conforming to DICOM standard PS 3. 10 is known as a DICOM files have two components: Header Contains information, organized in attributes or data elements, describing the contents of the file. Data Contains image pixel data, the structure and encoding of which is described in the header. The file can contain multiple frames as well as compressed data.

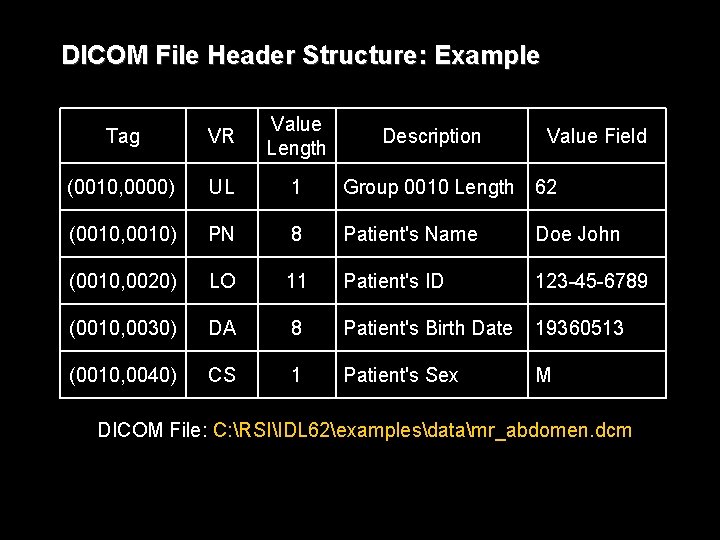
DICOM File Header Structure An attribute or data element is comprised of four fields: Tag VR Value Length Value Field • Tag - a unique numeric label that identifies the attribute • Value representation (VR) - a two-character code describing the type and format of the attribute's value • Value length - the length of the attribute's data • Value field - the attribute's data

DICOM File Header Structure • Attribute tags are expressed in the form (gggg, eeee) where gggg is the four-digit group and eeee is the four-digit element number, both in hexadecimal notation. • Even-valued groups are public, defined by the DICOM standard; odd-valued groups are private, defined by users of the format. Examples: – group 0008 : file creation information – group 0010 : patient information – group 0018 : imaging device parameters • A full list of attribute tags and their VRs are given in DICOM PS 3. 6, the Data Dictionary. A subset of these tags are listed in the Medical Imaging in IDL reference document.

DICOM File Header Structure: Example Tag VR Value Length (0010, 0000) UL 1 Group 0010 Length 62 (0010, 0010) PN 8 Patient's Name Doe John (0010, 0020) LO 11 Patient's ID 123 -45 -6789 (0010, 0030) DA 8 Patient's Birth Date 19360513 (0010, 0040) CS 1 Patient's Sex M Description Value Field DICOM File: C: RSIIDL 62examplesdatamr_abdomen. dcm

The IDL DICOM Toolkit The IDL DICOM toolkit consists of two interfaces to DICOM format files: IDLff. DICOMex These interfaces – conform to DICOM standard PS 3. 10 for reading DICOM files; IDLff. DICOMex can also write DICOM files – employ an object-oriented programming methodology – can be used at the command line or within an IDL program – are fully documented, with examples, in IDL's Online Help system

The IDL DICOM Toolkit IDLff. DICOM • provides basic read access to a DICOM file, including – group/element tag – value representation – length – data values – file header preamble – data dictionary description for individual elements – embedded sequences of elements • is included in the standard IDL distribution • has procedural wrappers QUERY_DICOM and READ_DICOM

The IDL DICOM Toolkit IDLff. DICOMex • can query, read, clone, or create new DICOM files • can read and write both public and private attributes including sequences and sets of repeating tags within sequences • can read and write compressed DICOM files on Windows and UNIX-based platforms • can copy DICOM attributes from one file to another • supports JPEG and JPEG 2000 files (except Mac OS X) • is built on an industry-standard commercial DICOM library from Merge Technologies, Inc. (www. merge. com) • requires a special IDL license

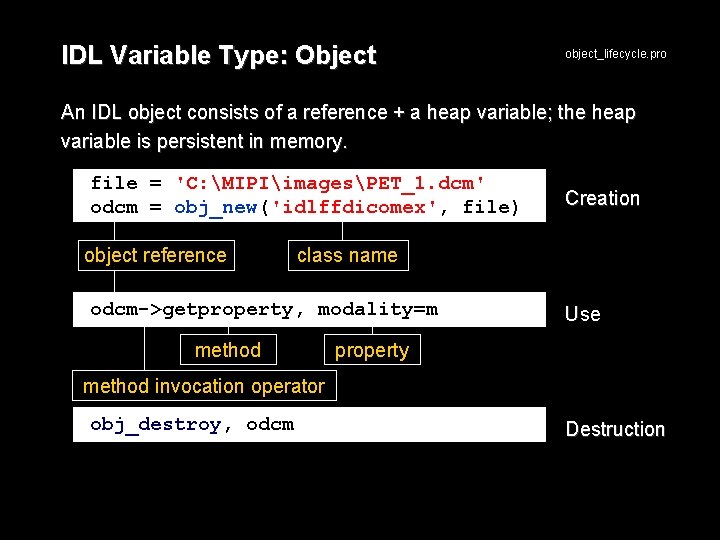
IDL Variable Type: Object IDL's DICOM toolkit is implemented using object technology, a programming methodology used, for example, in java and C++ Terminology: class - an abstract data type, where methods and properties are defined object - an instance of a class method - a program defining what an object can do property - a datum of an object Example: IDL is a programming language.

IDL Variable Type: Object object_lifecycle. pro An IDL object consists of a reference + a heap variable; the heap variable is persistent in memory. file = 'C: MIPIimagesPET_1. dcm' odcm = obj_new('idlffdicomex', file) object reference Creation class name odcm->getproperty, modality=m method Use property method invocation operator obj_destroy, odcm Destruction

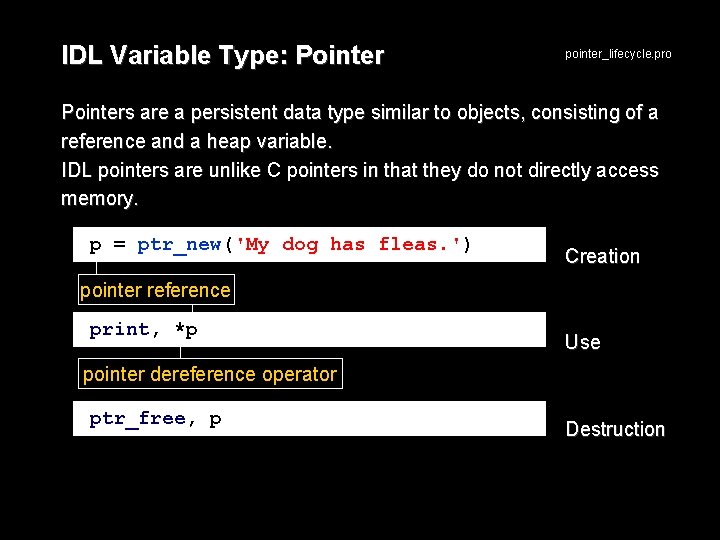
IDL Variable Type: Pointer pointer_lifecycle. pro Pointers are a persistent data type similar to objects, consisting of a reference and a heap variable. IDL pointers are unlike C pointers in that they do not directly access memory. p = ptr_new('My dog has fleas. ') Creation pointer reference print, *p Use pointer dereference operator ptr_free, p Destruction

The IDLff. DICOM Class • • Introduced in IDL 5. 2 (1998) to support the growing use of the DICOM standard. API odcm = obj_new('idlffdicom') • • Examples 1. dumping header tags to screen or file 2. retrieving header tag descriptions, VRs and values 3. reading image pixel data 4. data visualization (TV & TVSCL, IIMAGE, IVOLUME) Procedural wrappers

The IDLff. DICOM Class: Example 1 Read a DICOM file and dump its attributes to a text file. ; Make a new DICOM file object. odcm = obj_new('idlffdicom') ; Read the contents of a DICOM file = 'C: MIPIimagesCT_1. dcm' info = odcm->read(file) ; Dump its attributes to a text file. odcm->dumpelements, 'CT_1_hdr. txt' View the dump file CT_1_hdr. txt. idlffdicom_ex 1. pro

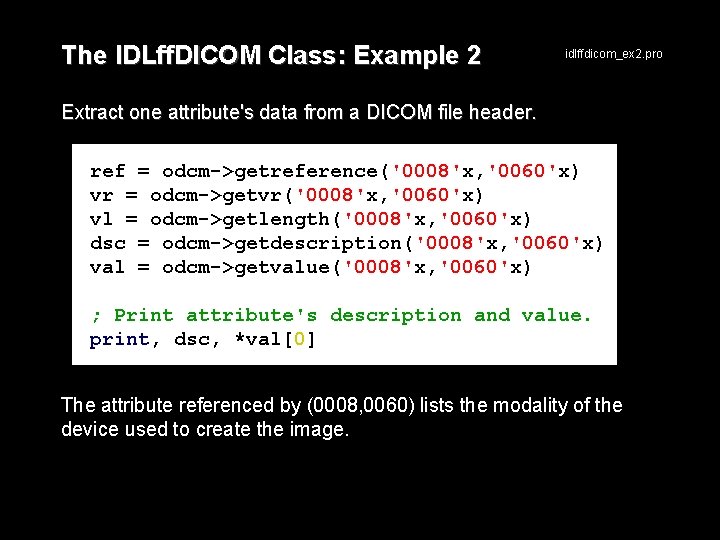
The IDLff. DICOM Class: Example 2 idlffdicom_ex 2. pro Extract one attribute's data from a DICOM file header. ref = odcm->getreference('0008'x, '0060'x) vr = odcm->getvr('0008'x, '0060'x) vl = odcm->getlength('0008'x, '0060'x) dsc = odcm->getdescription('0008'x, '0060'x) val = odcm->getvalue('0008'x, '0060'x) ; Print attribute's description and value. print, dsc, *val[0] The attribute referenced by (0008, 0060) lists the modality of the device used to create the image.

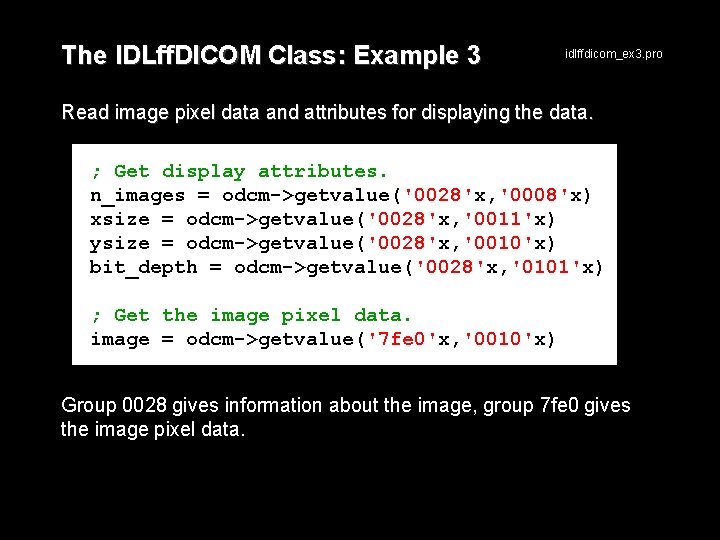
The IDLff. DICOM Class: Example 3 idlffdicom_ex 3. pro Read image pixel data and attributes for displaying the data. ; Get display attributes. n_images = odcm->getvalue('0028'x, '0008'x) xsize = odcm->getvalue('0028'x, '0011'x) ysize = odcm->getvalue('0028'x, '0010'x) bit_depth = odcm->getvalue('0028'x, '0101'x) ; Get the image pixel data. image = odcm->getvalue('7 fe 0'x, '0010'x) Group 0028 gives information about the image, group 7 fe 0 gives the image pixel data.

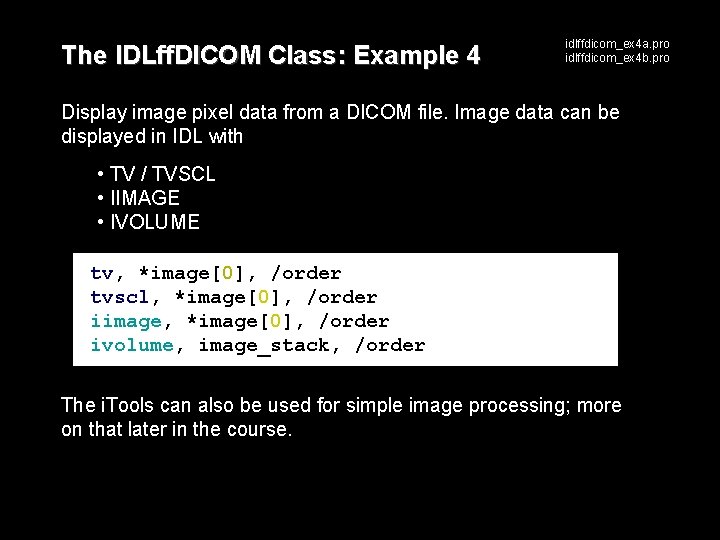
The IDLff. DICOM Class: Example 4 idlffdicom_ex 4 a. pro idlffdicom_ex 4 b. pro Display image pixel data from a DICOM file. Image data can be displayed in IDL with • TV / TVSCL • IIMAGE • IVOLUME tv, *image[0], /order tvscl, *image[0], /order iimage, *image[0], /order ivolume, image_stack, /order The i. Tools can also be used for simple image processing; more on that later in the course.

The IDLff. DICOM Class idlffdicom_wrappers. pro The IDLff. DICOM class has two procedural wrappers. QUERY_DICOM - tests that the file is a DICOM file, optionally returning a structure of information ok = query_dicom(dicom_file, info) READ_DICOM - reads the image pixel data from a DICOM file image = read_dicom(dicom_file) The wrappers are written in the IDL language.

The IDLff. DICOM Class: Problems 1. What is the patient's name in CT_2. dcm in the course files? What is the referring physician's name? 2. Read the image pixel data from mr_brain. dcm in the examples/data subdirectory of the IDL distribution. Display the data with TV in a window sized for the image. 3. Write a program to read the first four images from mr_abdomen. dcm (also in examples/data) and display them in a 2 x 2 grid in IIMAGE.

The IDLff. DICOMex Class • Introduced in IDL 6. 1 (2004) to upgrade IDL's ability to work with DICOM, especially the ability to clone and write DICOM files. • API odcm = obj_new('idlffdicomex', file) • Examples – read the attributes into an array of IDL structures – retrieve and display image pixel data – query the attributes of a DICOM file – clone a DICOM file and alter its attributes – create a new DICOM file • The DICOM_VIEWER demo application

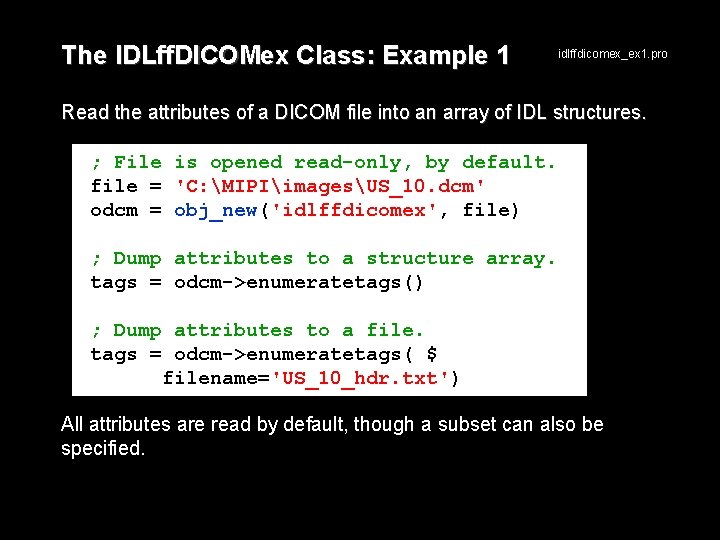
The IDLff. DICOMex Class: Example 1 idlffdicomex_ex 1. pro Read the attributes of a DICOM file into an array of IDL structures. ; File is opened read-only, by default. file = 'C: MIPIimagesUS_10. dcm' odcm = obj_new('idlffdicomex', file) ; Dump attributes to a structure array. tags = odcm->enumeratetags() ; Dump attributes to a file. tags = odcm->enumeratetags( $ filename='US_10_hdr. txt') All attributes are read by default, though a subset can also be specified.

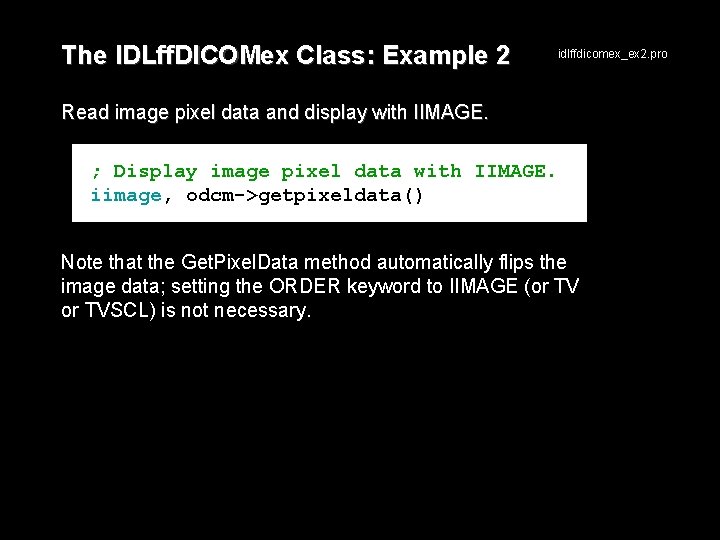
The IDLff. DICOMex Class: Example 2 idlffdicomex_ex 2. pro Read image pixel data and display with IIMAGE. ; Display image pixel data with IIMAGE. iimage, odcm->getpixeldata() Note that the Get. Pixel. Data method automatically flips the image data; setting the ORDER keyword to IIMAGE (or TVSCL) is not necessary.

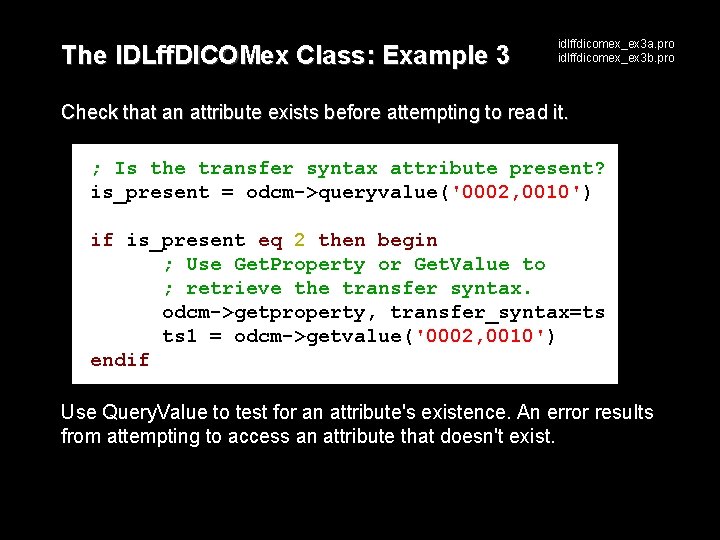
The IDLff. DICOMex Class: Example 3 idlffdicomex_ex 3 a. pro idlffdicomex_ex 3 b. pro Check that an attribute exists before attempting to read it. ; Is the transfer syntax attribute present? is_present = odcm->queryvalue('0002, 0010') if is_present eq 2 then begin ; Use Get. Property or Get. Value to ; retrieve the transfer syntax. odcm->getproperty, transfer_syntax=ts ts 1 = odcm->getvalue('0002, 0010') endif Use Query. Value to test for an attribute's existence. An error results from attempting to access an attribute that doesn't exist.

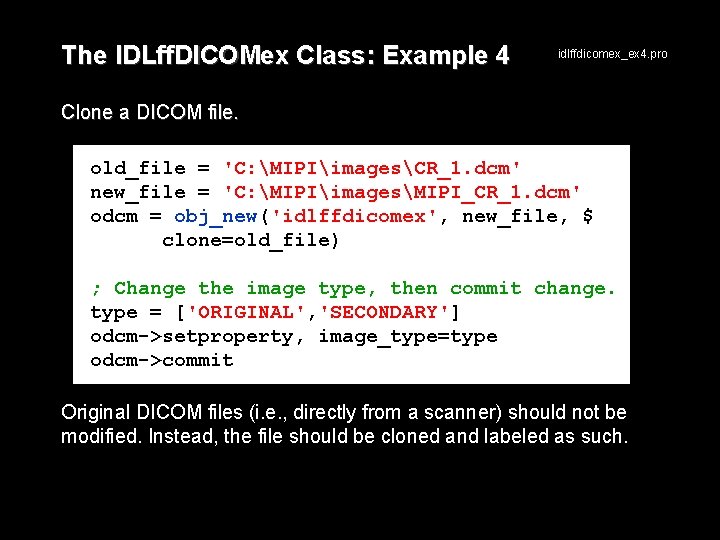
The IDLff. DICOMex Class: Example 4 idlffdicomex_ex 4. pro Clone a DICOM file. old_file = 'C: MIPIimagesCR_1. dcm' new_file = 'C: MIPIimagesMIPI_CR_1. dcm' odcm = obj_new('idlffdicomex', new_file, $ clone=old_file) ; Change the image type, then commit change. type = ['ORIGINAL', 'SECONDARY'] odcm->setproperty, image_type=type odcm->commit Original DICOM files (i. e. , directly from a scanner) should not be modified. Instead, the file should be cloned and labeled as such.

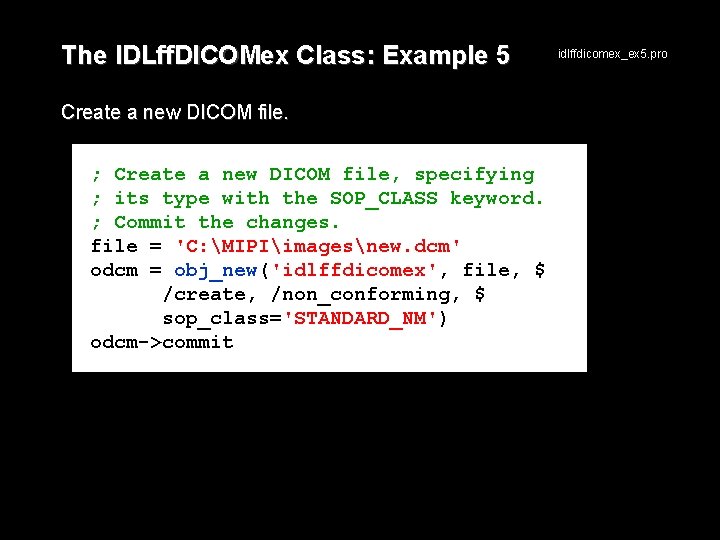
The IDLff. DICOMex Class: Example 5 Create a new DICOM file. ; Create a new DICOM file, specifying ; its type with the SOP_CLASS keyword. ; Commit the changes. file = 'C: MIPIimagesnew. dcm' odcm = obj_new('idlffdicomex', file, $ /create, /non_conforming, $ sop_class='STANDARD_NM') odcm->commit idlffdicomex_ex 5. pro

The IDLff. DICOMex Class: Problems 1. Read only the tags from group (0002) in the clone file MIPI_CR_1. img. Write the tags to a text file. Compare them with the elements of the same group in CR_1. dcm. 2. Write a program to read the image pixel data from mr_brain. dcm (in the examples/data subdirectory of the IDL distribution) and display them with TV in a window sized for the image. Compare with problem #2 for IDLff. DICOM. 3. Clone CR_1. dcm and reduce the size of the file by selecting a transfer syntax that supports compression. 4. Read the file ctscan. dat from the examples/data subdirectory and create a new (nonconforming) DICOM file with its image pixel data.

Further Information For more information on IDL's DICOM capabilites, see • http: //www. rsinc. com/dicom – Toolkit information – Conformance statement • http: //www. rsinc. com/webinar – "IDL Medical Imaging Suite" presentation • Medical Imaging in IDL in the IDL Online Help

Part II: Image Processing in IDL

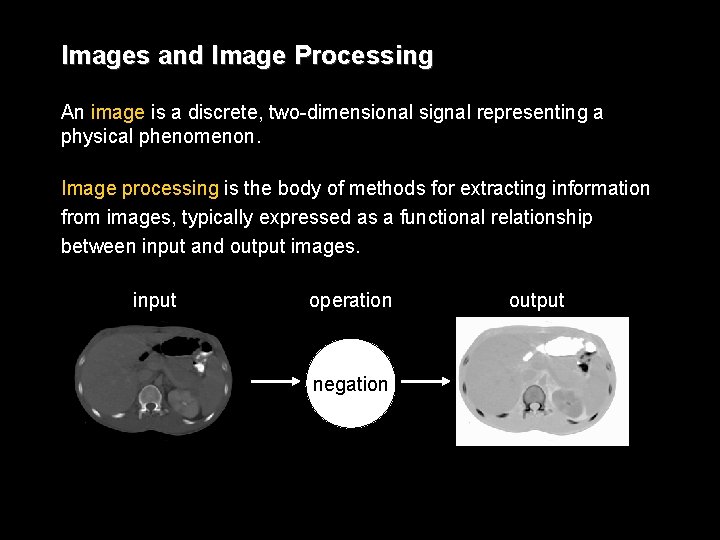
Images and Image Processing An image is a discrete, two-dimensional signal representing a physical phenomenon. Image processing is the body of methods for extracting information from images, typically expressed as a functional relationship between input and output images. input operation negation output

Outline 1. IDL arrays and diagnostic/statistical routines 2. Pixel operations 1. stretching/scaling, window center and width, thresholding, masking, negation, mathematical operations, equalization 3. Neighborhood operations 1. smoothing, median filtering, convolution, sharpening, edge detection 4. Global transforms 1. Fourier and Radon transforms 5. Geometric operations 1. rotating, scaling and resizing images; registration, fusion 6. Feature extraction methods 1. regions of interest (ROI), region growing, morphological operators, segmentation

IDL Arrays and Diagnostic / Statistical Routines

IDL Arrays array_operation_ex. pro • IDL is an array-based language. • Images, image stacks and cines are represented in IDL as 2 -, 3 or 4 -D arrays, consisting of – 1 -, 2 -, 4 - or 8 -byte unsigned integers – 2 -, 4 - or 8 -byte signed integers – 4 - or 8 -byte floating point values • Array operations are faster than scalar operations. – example: find maximum pixel value of image • IDL has many built-in, array-oriented routines for image processing (see Online Help).

Diagnostic and Statistical Routines helpful_routines_ex. pro A list of routines used to get information about an image. HELP SIZE MIN MAX WHERE ARRAY_INDICES MOMENT MEDIAN ARRAY_EQUAL TOTAL PRODUCT IMAGE_STATISTICS FINITE HISTOGRAM N_ELEMENTS UNIQ SORT

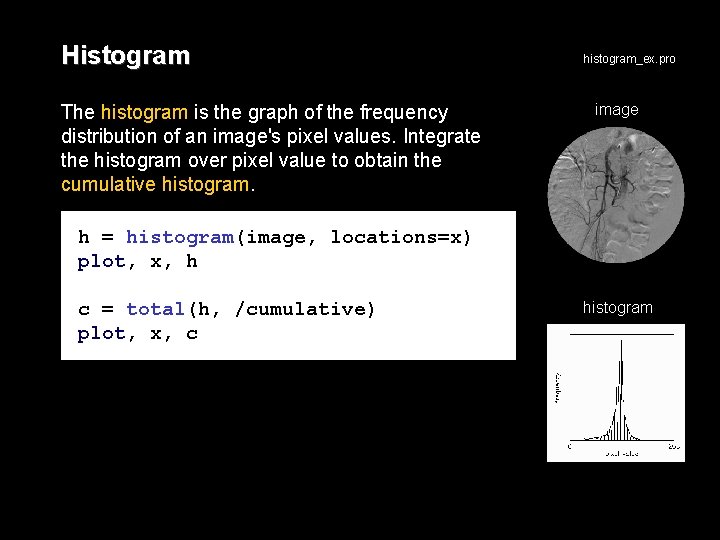
Histogram The histogram is the graph of the frequency distribution of an image's pixel values. Integrate the histogram over pixel value to obtain the cumulative histogram_ex. pro image h = histogram(image, locations=x) plot, x, h c = total(h, /cumulative) plot, x, c histogram

Problems 1. Write a function that returns the range of values in an array. 2. Write a function that returns the odd values of an input array. (Hint: use WHERE)

Pixel Operations

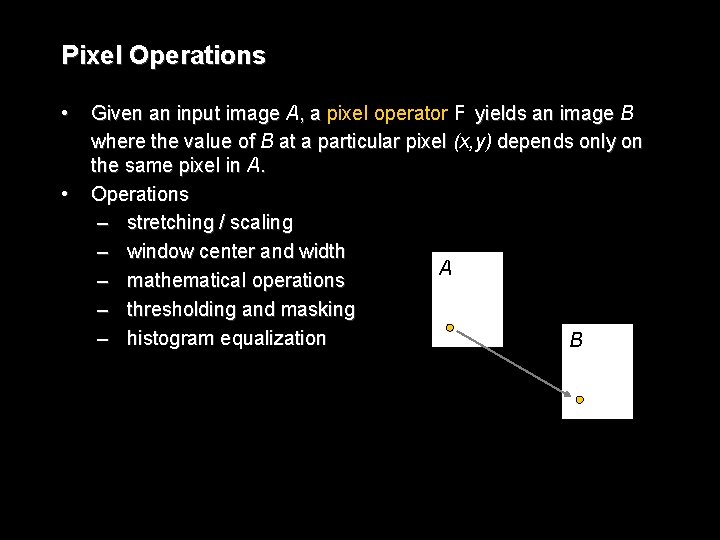
Pixel Operations • • Given an input image A, a pixel operator F yields an image B where the value of B at a particular pixel (x, y) depends only on the same pixel in A. Operations – stretching / scaling – window center and width A – mathematical operations – thresholding and masking – histogram equalization B

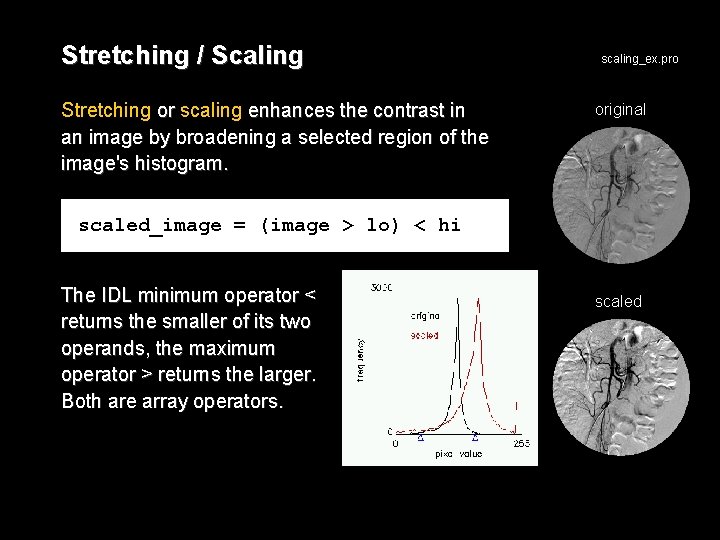
Stretching / Scaling Stretching or scaling enhances the contrast in an image by broadening a selected region of the image's histogram. scaling_ex. pro original scaled_image = (image > lo) < hi The IDL minimum operator < returns the smaller of its two operands, the maximum operator > returns the larger. Both are array operators. scaled

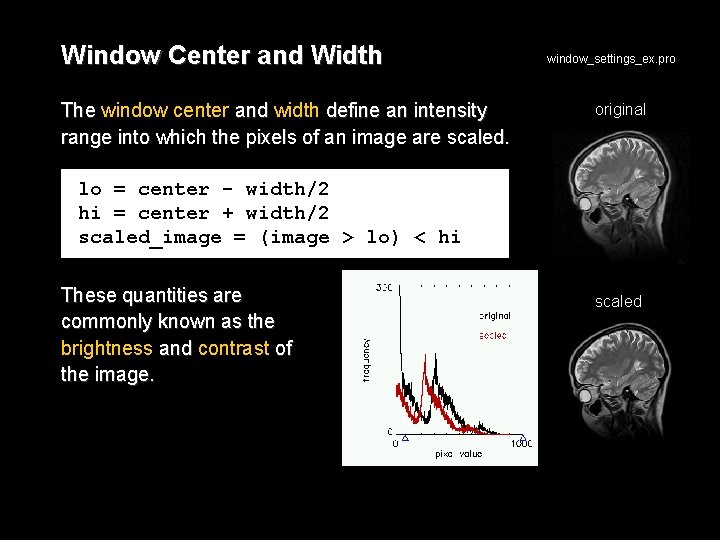
Window Center and Width The window center and width define an intensity range into which the pixels of an image are scaled. window_settings_ex. pro original lo = center - width/2 hi = center + width/2 scaled_image = (image > lo) < hi These quantities are commonly known as the brightness and contrast of the image. scaled

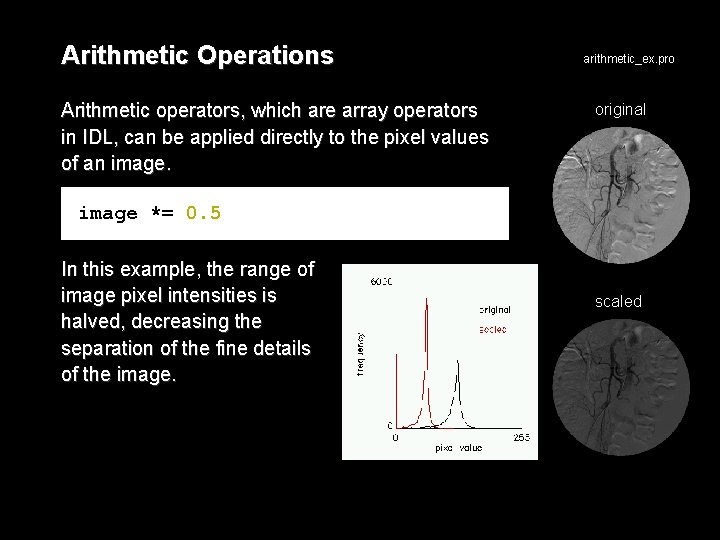
Arithmetic Operations Arithmetic operators, which are array operators in IDL, can be applied directly to the pixel values of an image. arithmetic_ex. pro original image *= 0. 5 In this example, the range of image pixel intensities is halved, decreasing the separation of the fine details of the image. scaled

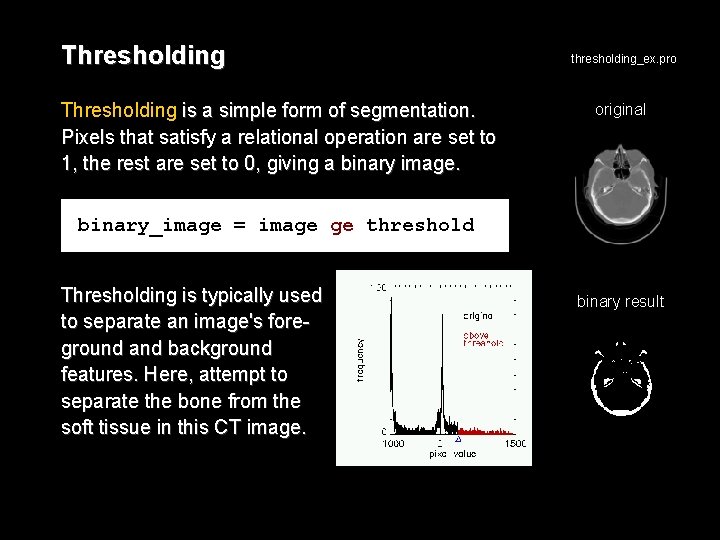
Thresholding is a simple form of segmentation. Pixels that satisfy a relational operation are set to 1, the rest are set to 0, giving a binary image. thresholding_ex. pro original binary_image = image ge threshold Thresholding is typically used to separate an image's foreground and background features. Here, attempt to separate the bone from the soft tissue in this CT image. binary result

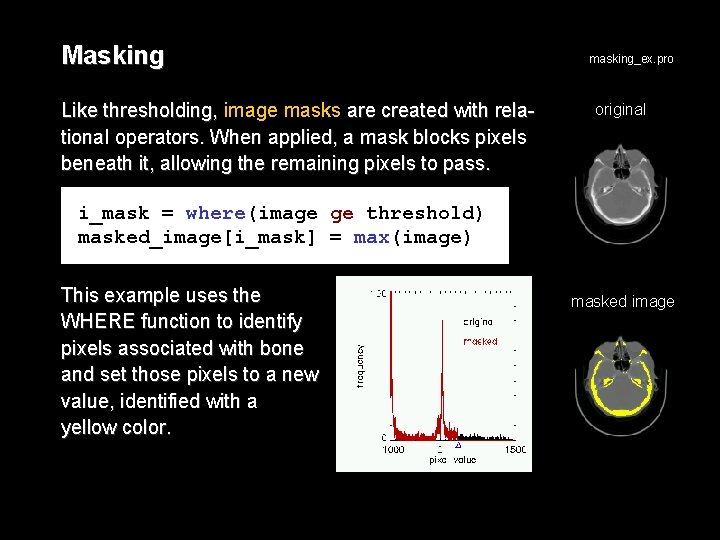
Masking Like thresholding, image masks are created with relational operators. When applied, a mask blocks pixels beneath it, allowing the remaining pixels to pass. masking_ex. pro original i_mask = where(image ge threshold) masked_image[i_mask] = max(image) This example uses the WHERE function to identify pixels associated with bone and set those pixels to a new value, identified with a yellow color. masked image

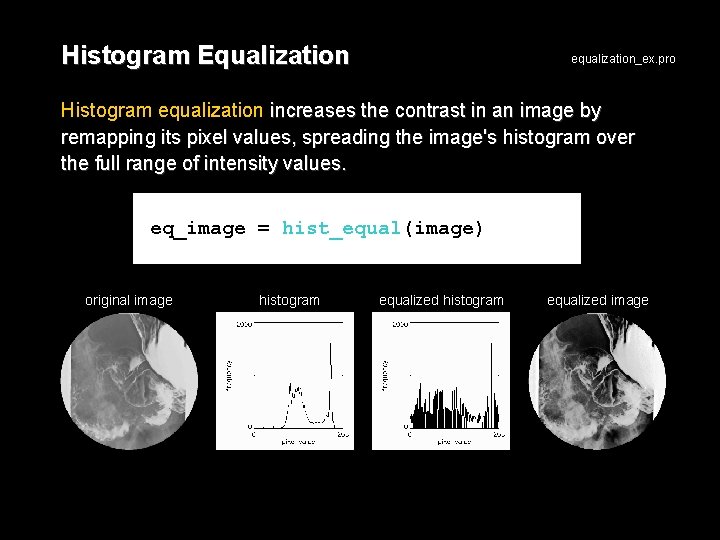
Histogram Equalization equalization_ex. pro Histogram equalization increases the contrast in an image by remapping its pixel values, spreading the image's histogram over the full range of intensity values. eq_image = hist_equal(image) original image histogram equalized image

Problems 1. Make an IDL function to set the window center and width for an image. 2. Construct a mask to remove a square region of 100 x 100 pixels from the center of the image kidney 20. dcm. 3. Create a new image from kidney 20. dcm where the black pixels around the edge are transparent.

Neighborhood Operations

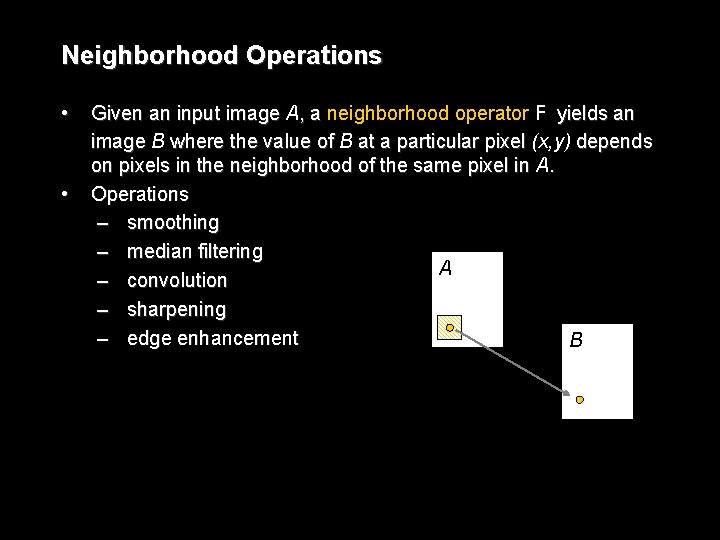
Neighborhood Operations • • Given an input image A, a neighborhood operator F yields an image B where the value of B at a particular pixel (x, y) depends on pixels in the neighborhood of the same pixel in A. Operations – smoothing – median filtering A – convolution – sharpening – edge enhancement B

Smoothing smoothing_ex. pro Smoothing removes high-frequency information from an image. Smoothing is typically used to attenuate noise or soften an image. original smoothed_image = smooth(image, 5) The SMOOTH function is used to apply a rectangular running-mean (or tophat) filter to an image. smoothing kernel filtered image

Median Filtering Median filtering replaces each point in an image with the median intensity value of a twodimensional neighborhood of a given width. median_filtering_ex. pro original smoothed_image = median(image, 5) Median filtering removes outlying values from an image without altering or introducing new pixels into the image, thereby preserving edges in the image. filtered image

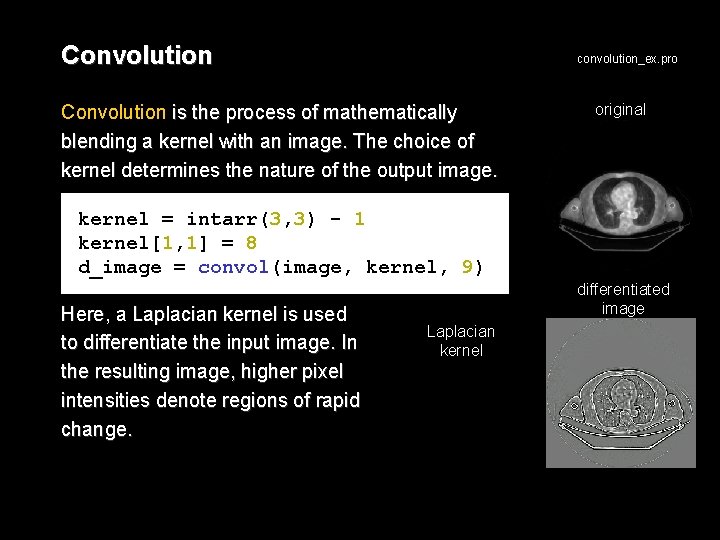
Convolution convolution_ex. pro Convolution is the process of mathematically blending a kernel with an image. The choice of kernel determines the nature of the output image. original kernel = intarr(3, 3) - 1 kernel[1, 1] = 8 d_image = convol(image, kernel, 9) Here, a Laplacian kernel is used to differentiate the input image. In the resulting image, higher pixel intensities denote regions of rapid change. differentiated image Laplacian kernel

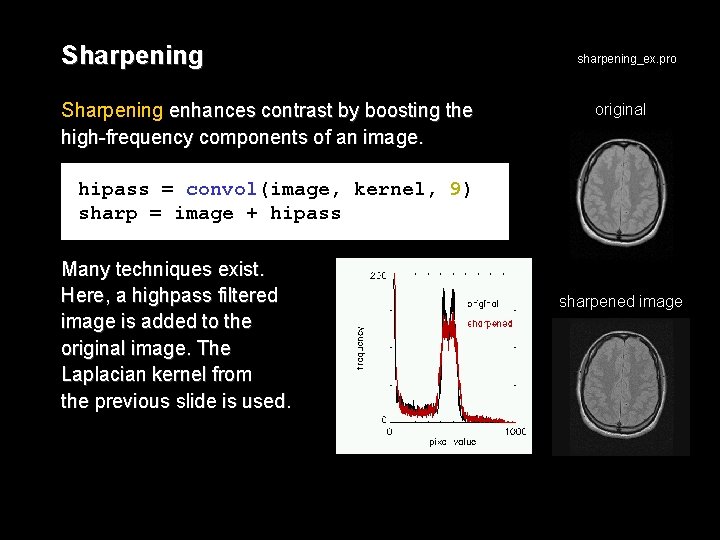
Sharpening enhances contrast by boosting the high-frequency components of an image. sharpening_ex. pro original hipass = convol(image, kernel, 9) sharp = image + hipass Many techniques exist. Here, a highpass filtered image is added to the original image. The Laplacian kernel from the previous slide is used. sharpened image

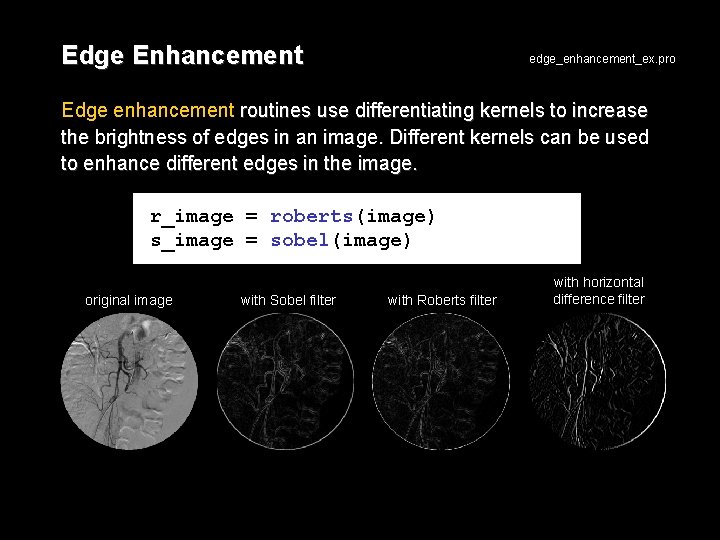
Edge Enhancement edge_enhancement_ex. pro Edge enhancement routines use differentiating kernels to increase the brightness of edges in an image. Different kernels can be used to enhance different edges in the image. r_image = roberts(image) s_image = sobel(image) original image with Sobel filter with Roberts filter with horizontal difference filter

Problems 1. Write a program to display the difference between images filtered with the SMOOTH and MEDIAN functions. 2. Use CONVOL to apply a Gaussian smoothing kernel to an image. Compare the result with output from SMOOTH for the same kernel size. 3. Demonstrate that prior smoothing of an image can improve the results of edge enhancement routines.

Global Transforms

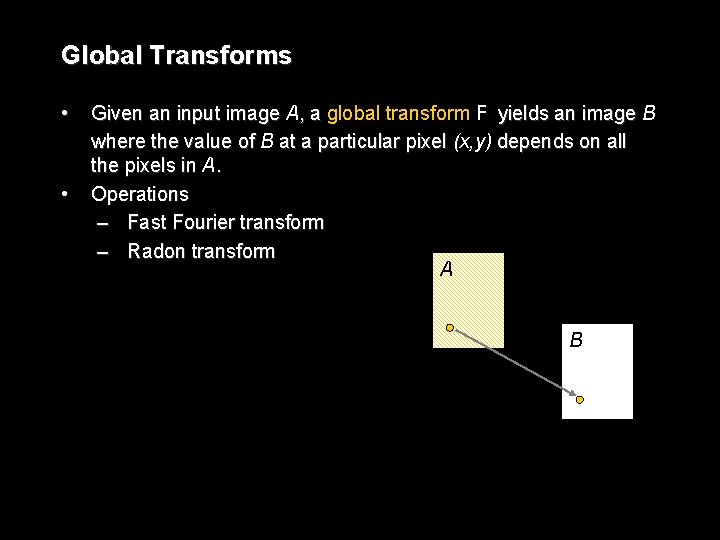
Global Transforms • • Given an input image A, a global transform F yields an image B where the value of B at a particular pixel (x, y) depends on all the pixels in A. Operations – Fast Fourier transform – Radon transform A B

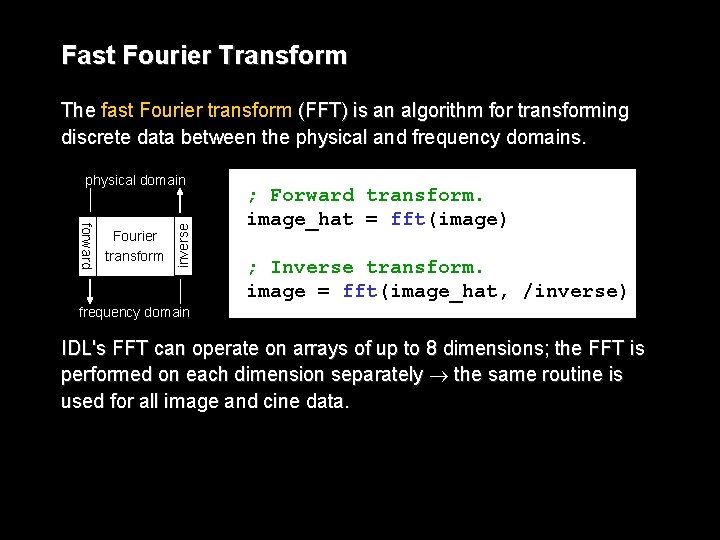
Fast Fourier Transform The fast Fourier transform (FFT) is an algorithm for transforming discrete data between the physical and frequency domains. forward Fourier transform inverse physical domain ; Forward transform. image_hat = fft(image) ; Inverse transform. image = fft(image_hat, /inverse) frequency domain IDL's FFT can operate on arrays of up to 8 dimensions; the FFT is performed on each dimension separately the same routine is used for all image and cine data.

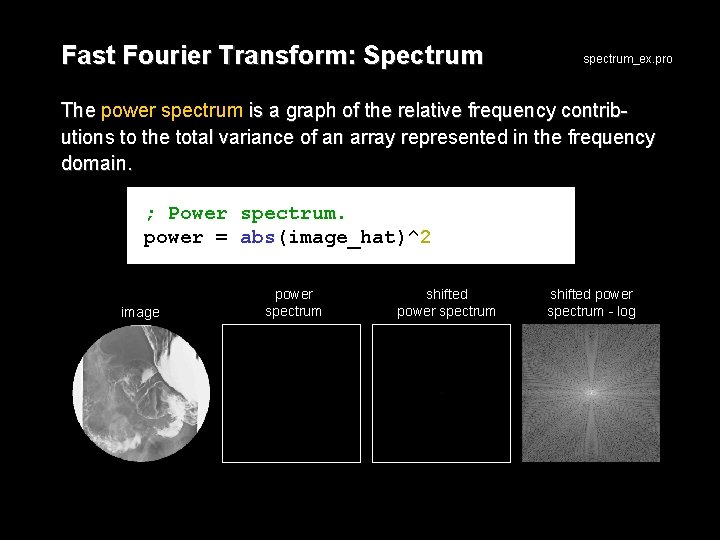
Fast Fourier Transform: Spectrum spectrum_ex. pro The power spectrum is a graph of the relative frequency contributions to the total variance of an array represented in the frequency domain. ; Power spectrum. power = abs(image_hat)^2 image power spectrum shifted power spectrum - log

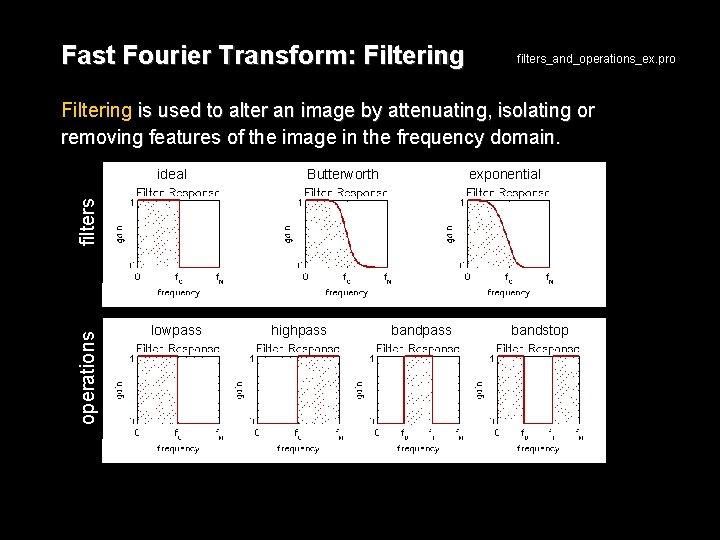
Fast Fourier Transform: Filtering filters_and_operations_ex. pro Filtering is used to alter an image by attenuating, isolating or removing features of the image in the frequency domain. Butterworth exponential operations filters ideal lowpass highpass bandstop

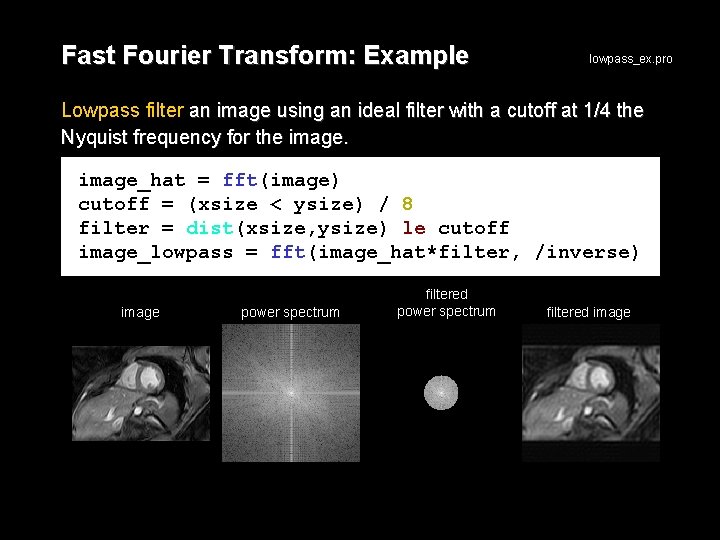
Fast Fourier Transform: Example lowpass_ex. pro Lowpass filter an image using an ideal filter with a cutoff at 1/4 the Nyquist frequency for the image_hat = fft(image) cutoff = (xsize < ysize) / 8 filter = dist(xsize, ysize) le cutoff image_lowpass = fft(image_hat*filter, /inverse) image power spectrum filtered image

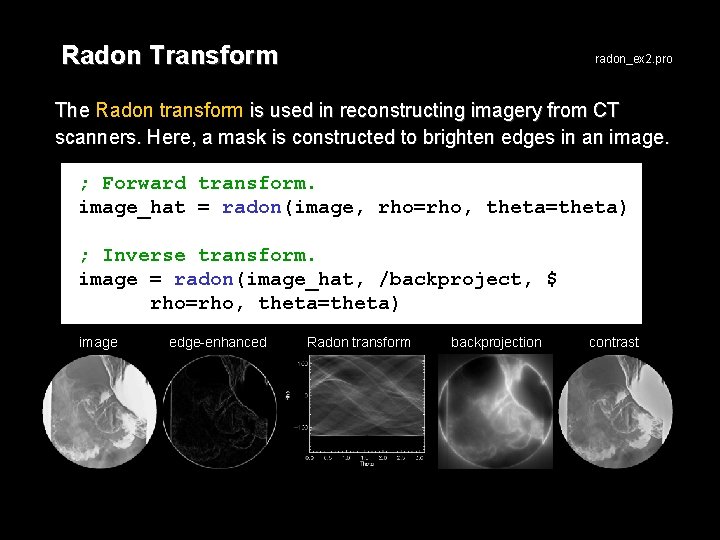
Radon Transform radon_ex 2. pro The Radon transform is used in reconstructing imagery from CT scanners. Here, a mask is constructed to brighten edges in an image. ; Forward transform. image_hat = radon(image, rho=rho, theta=theta) ; Inverse transform. image = radon(image_hat, /backproject, $ rho=rho, theta=theta) image edge-enhanced Radon transform backprojection contrast

Problems 1. Sharpen an image using Fourier filtering. Compare the results with those from SHARPENING_EX in the previous section.

Geometric Operations

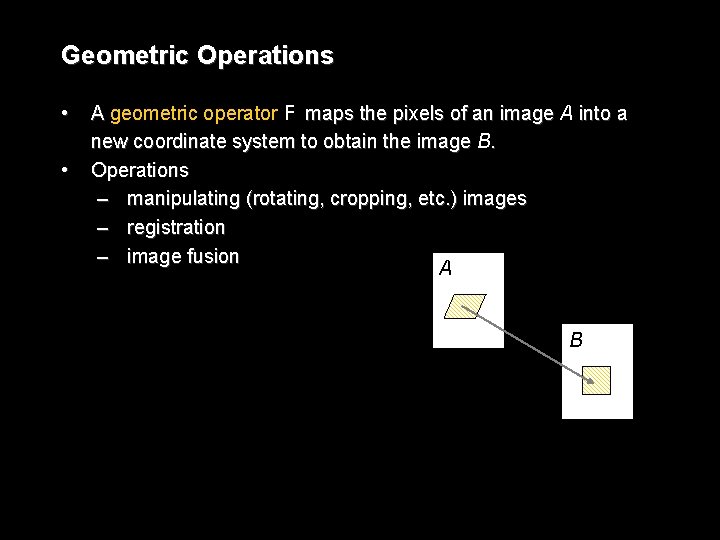
Geometric Operations • • A geometric operator F maps the pixels of an image A into a new coordinate system to obtain the image B. Operations – manipulating (rotating, cropping, etc. ) images – registration – image fusion A B

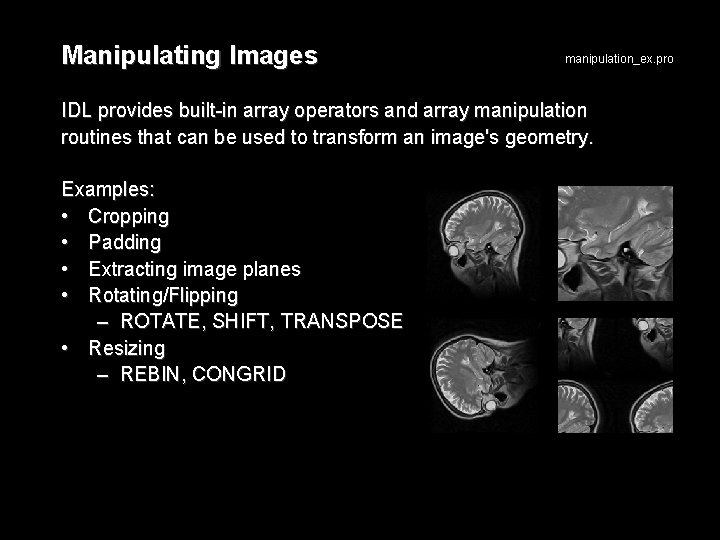
Manipulating Images manipulation_ex. pro IDL provides built-in array operators and array manipulation routines that can be used to transform an image's geometry. Examples: • Cropping • Padding • Extracting image planes • Rotating/Flipping – ROTATE, SHIFT, TRANSPOSE • Resizing – REBIN, CONGRID

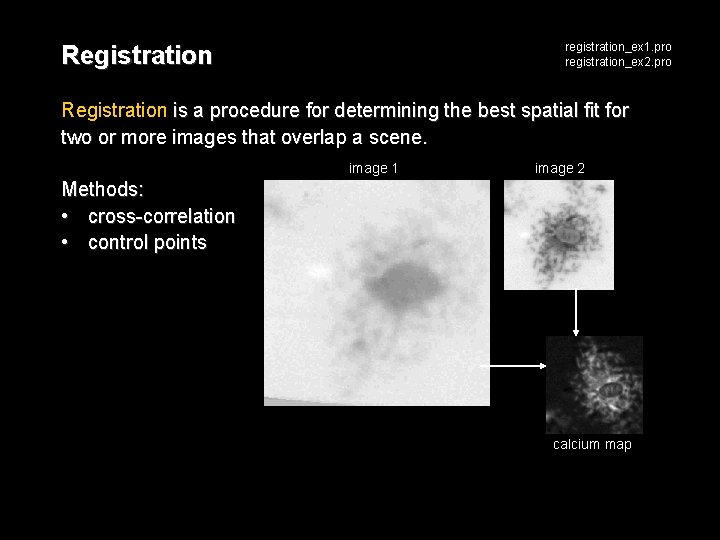
Registration registration_ex 1. pro registration_ex 2. pro Registration is a procedure for determining the best spatial fit for two or more images that overlap a scene. image 1 image 2 Methods: • cross-correlation • control points calcium map

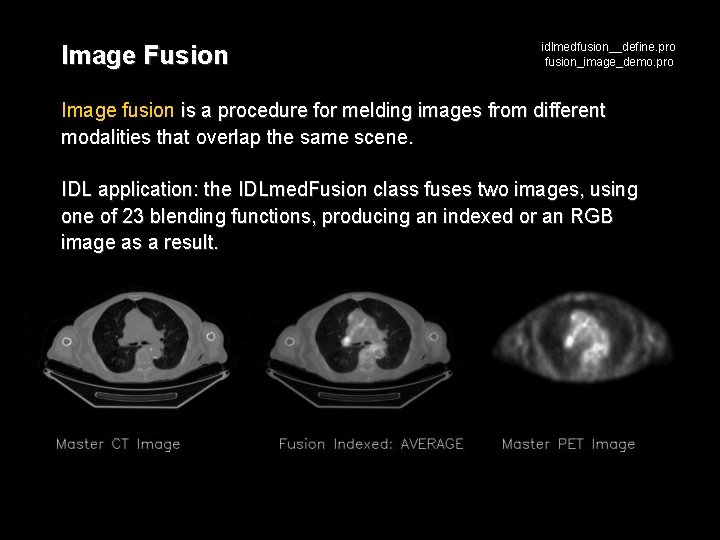
Image Fusion idlmedfusion__define. pro fusion_image_demo. pro Image fusion is a procedure for melding images from different modalities that overlap the same scene. IDL application: the IDLmed. Fusion class fuses two images, using one of 23 blending functions, producing an indexed or an RGB image as a result.

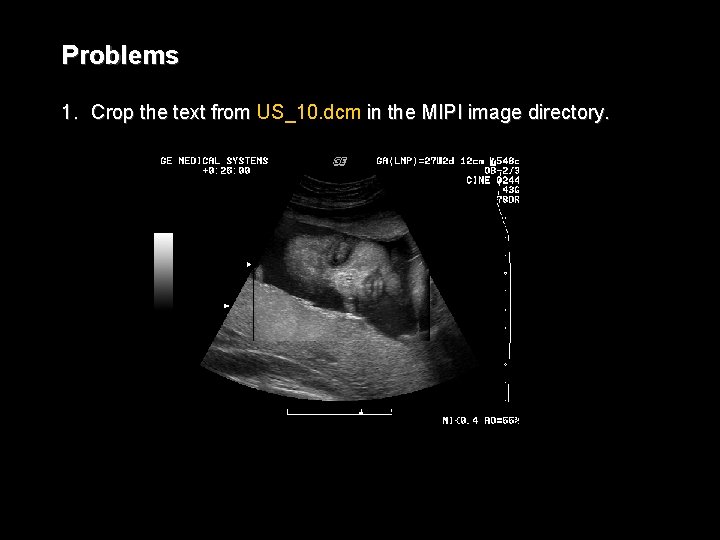
Problems 1. Crop the text from US_10. dcm in the MIPI image directory.

Feature Extraction Methods

Feature Extraction Methods • • Given an input image A, a feature extraction operator F selects, emphasizes, or modifies certain aspects of image A. Operations – regions of interest (ROIs), region growing – morphological operations – segmentation A B

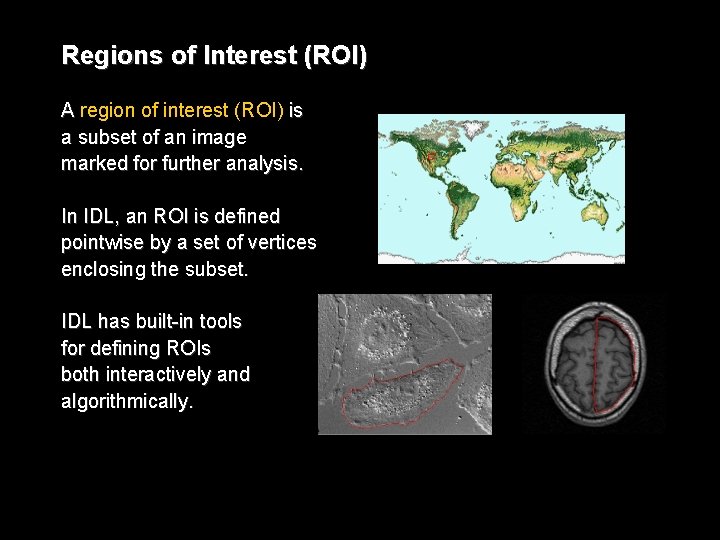
Regions of Interest (ROI) A region of interest (ROI) is a subset of an image marked for further analysis. In IDL, an ROI is defined pointwise by a set of vertices enclosing the subset. IDL has built-in tools for defining ROIs both interactively and algorithmically.

Regions of Interest (ROI) • Manual or interactive ROI definition: – XROI – IIMAGE • Automatic or algorithmic ROI definition: – CONTOUR – SEARCH 2 D / SEARCH 3 D – REGION_GROW • Quantification: – IDLan. ROI class – IMAGE_STATISTICS

Regions of Interest (ROI) • An object of the IDLan. ROI class represents the set of vertices defining an ROI. • API oroi = obj_new('idlanroi') • Used for – computing statistics – adding/removing points – generating a binary mask – rotating, translating or scaling the ROI – checking whether a point is interior/exterior to the ROI

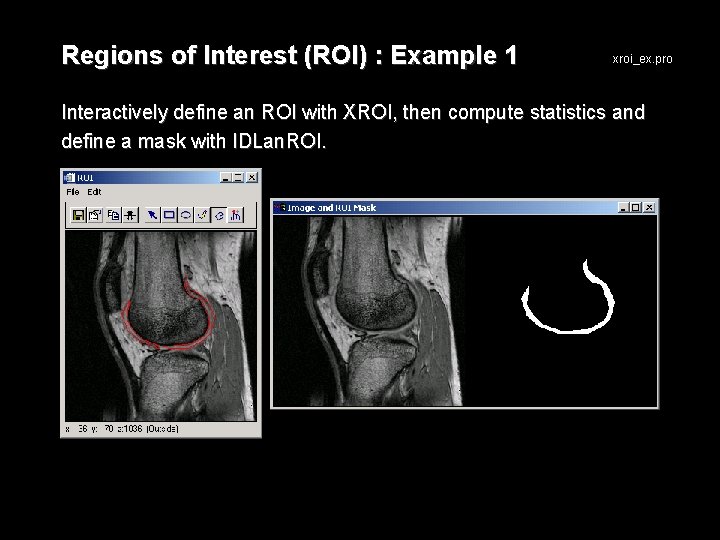
Regions of Interest (ROI) : Example 1 xroi_ex. pro Interactively define an ROI with XROI, then compute statistics and define a mask with IDLan. ROI.

Regions of Interest (ROI): Example 2 Perform a flood fill analysis with SEARCH 2 D, automatically defining a region. Recover the bounding points of the region with CONTOUR, obtaining the vertices of an ROI. Compute ROI statistics; construct an image mask. search 2 d_ex. pro

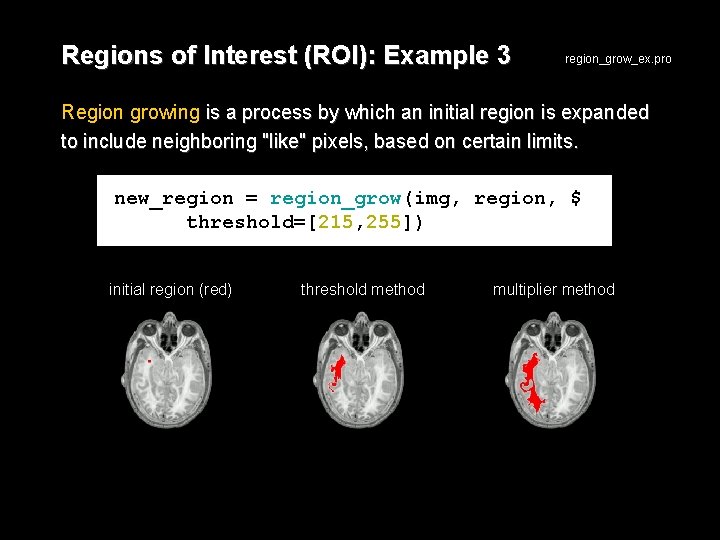
Regions of Interest (ROI): Example 3 region_grow_ex. pro Region growing is a process by which an initial region is expanded to include neighboring "like" pixels, based on certain limits. new_region = region_grow(img, region, $ threshold=[215, 255]) initial region (red) threshold method multiplier method

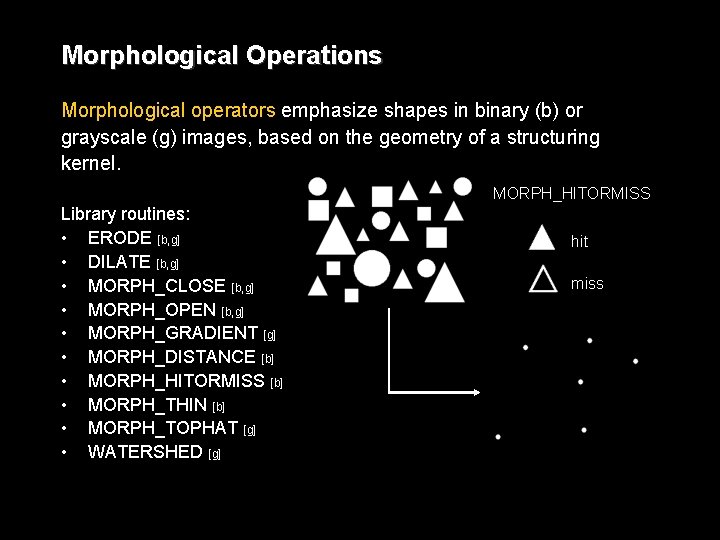
Morphological Operations Morphological operators emphasize shapes in binary (b) or grayscale (g) images, based on the geometry of a structuring kernel. MORPH_HITORMISS Library routines: • ERODE [b, g] • DILATE [b, g] • MORPH_CLOSE [b, g] • MORPH_OPEN [b, g] • MORPH_GRADIENT [g] • MORPH_DISTANCE [b] • MORPH_HITORMISS [b] • MORPH_THIN [b] • MORPH_TOPHAT [g] • WATERSHED [g] hit miss

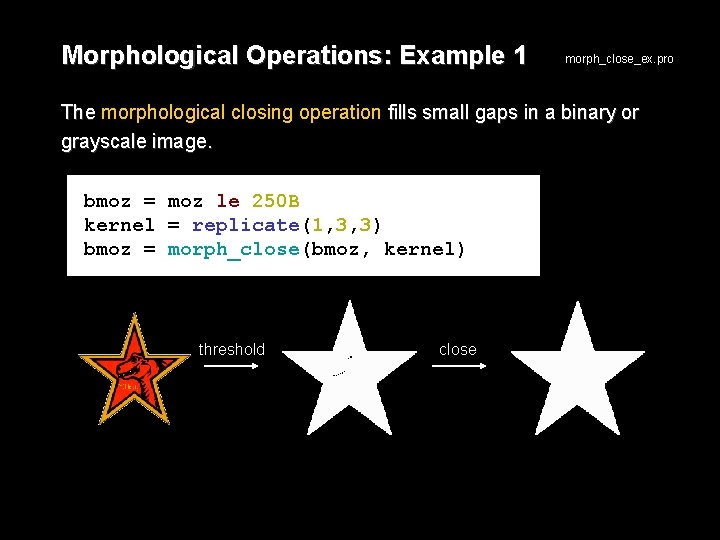
Morphological Operations: Example 1 morph_close_ex. pro The morphological closing operation fills small gaps in a binary or grayscale image. bmoz = moz le 250 B kernel = replicate(1, 3, 3) bmoz = morph_close(bmoz, kernel) threshold close

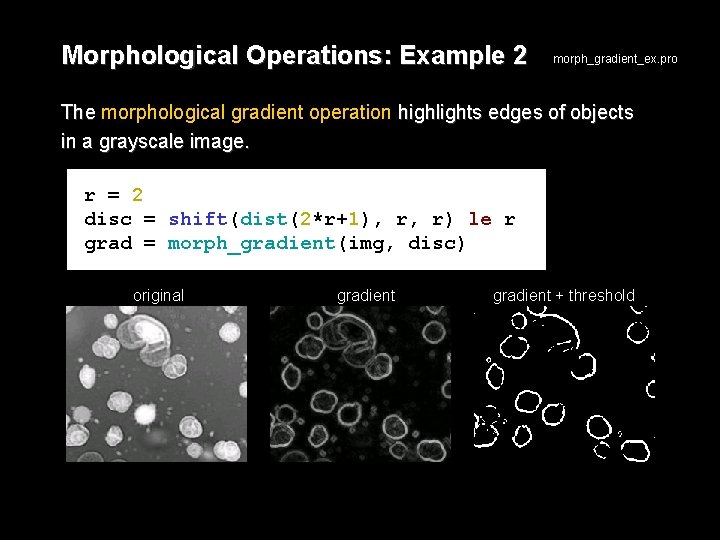
Morphological Operations: Example 2 morph_gradient_ex. pro The morphological gradient operation highlights edges of objects in a grayscale image. r = 2 disc = shift(dist(2*r+1), r, r) le r grad = morph_gradient(img, disc) original gradient + threshold

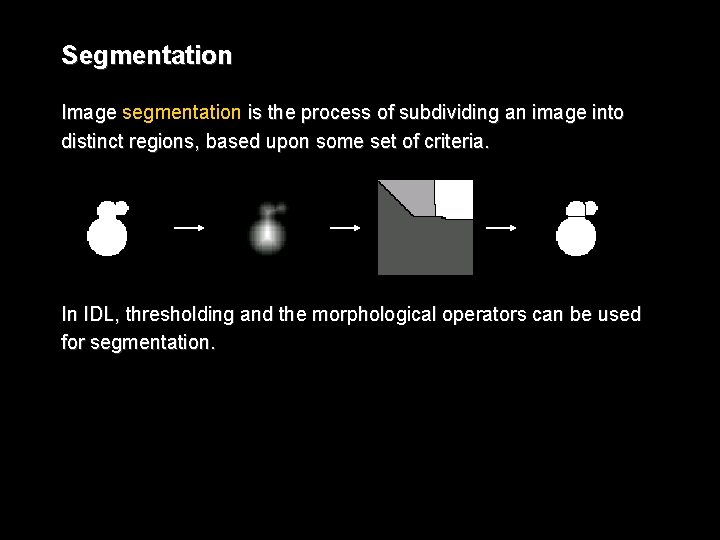
Segmentation Image segmentation is the process of subdividing an image into distinct regions, based upon some set of criteria. In IDL, thresholding and the morphological operators can be used for segmentation.

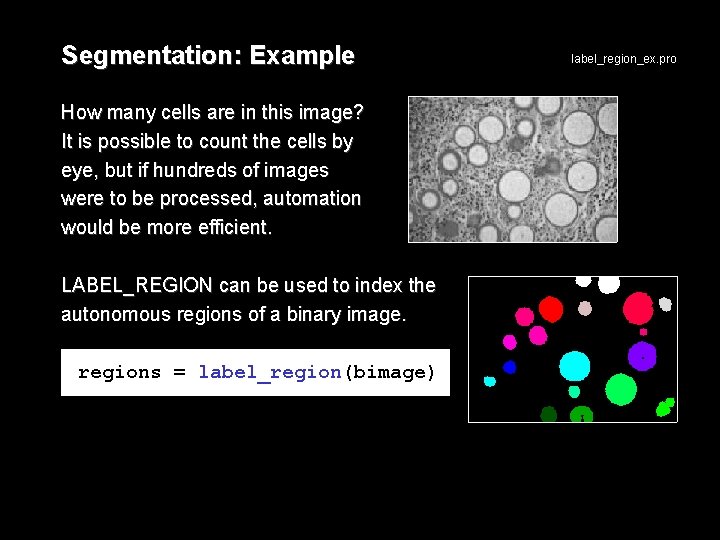
Segmentation: Example How many cells are in this image? It is possible to count the cells by eye, but if hundreds of images were to be processed, automation would be more efficient. LABEL_REGION can be used to index the autonomous regions of a binary image. regions = label_region(bimage) label_region_ex. pro

Problems 1. Define ROIs from the two regions created in the example program REGION_GROW_EX. 2. Experiment with altering the geometry of the structuring elements in the MORPH_* examples. How sensitive are the results to the size and shape of the structuring elements? 3. Use LABEL_REGION to count the cells in rbcells. jpg in the examples/data subdirectory.

References DICOM information • Official NEMA site http: //medical. nema. org • Prof. Chris Rorden http: //www. psychology. nottingham. ac. uk/staff/cr 1/dicom. html • Dr. David Clunie http: //www. dclunie. com/medical -image-faq/html/toc. html Image processing • Easton, R. Fundamentals of Digital Image Processing. Short course, Rochester Institute of Technology, July 1988. • Gonzales, R. C. and R. E. Woods. Digital Image Processing. Reading, Massachusetts: Addison-Wesley, 1992. • Russ, J. C. The Image Processing Handbook. Second edition. Boca Raton, Florida: CRC Press, 1995.

Contact Information RSI Global Services 4990 Pearl East Circle Boulder, CO 80301 tel fax +1 303 786 9900 +1 303 786 9909 services@rsinc. com training@rsinc. com http: //www. rsinc. com