Mechanisms Involved in Protein Synthesis Part 5 Codons

















- Slides: 19

Mechanisms Involved in Protein Synthesis Part 5: Codons and The Genetic Code

The Genetic Code • If the bases are counted in groups of three, 64 combinations are possible (43 = 64). • The three nucleotides which specify one amino acid are known as a codon. • 61 codons specify the 20 amino acids and 3 are stop codons: – UAA, UAG and UGA • The code is degenerate - 18 of the 20 amino acids are specified by multiple codons which are grouped together in the genetic code. • Codons that specify the same amino acid are called synonyms e. g. CAU and CAC are synonymous codons for histamine. In general, most synonyms differ only in the last base of the codon.

Figure 1. 20 Genomes 3 (© Garland Science 2007)

Figure 1. 21 Genomes 3 (© Garland Science 2007)

Table 1. 2 Genomes 3 (© Garland Science 2007)

Part 6 Translation

The three stages of protein synthesis are: 1. Initiation – assembly of a ribosome on an m. RNA 2. Elongation – repeated cycles of amino acid delivery and movement along the ribosome 3. Termination – release of the polypeptide chain

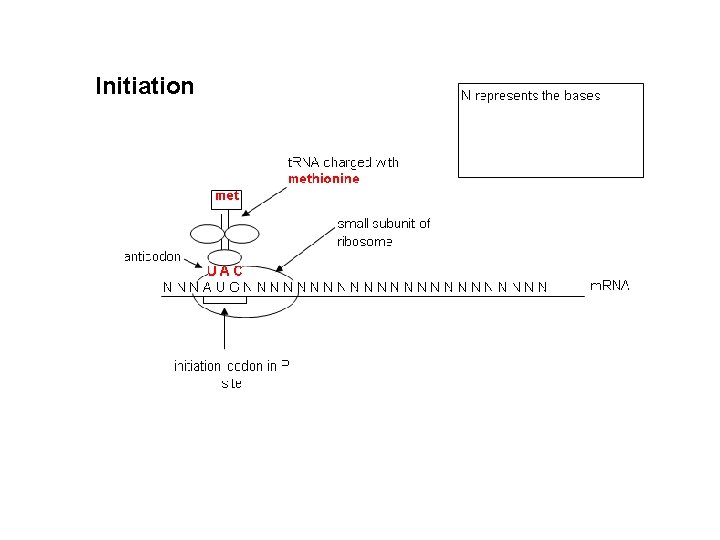
Initiation of Translation commences when a ribosome attaches to the m. RNA transcript. Ribosomes consist of two r. RNAs and are made up from two subunits, one small and one large, which form together around the m. RNA.

In eukaryotes: • Initiation involves the t. RNA binding to the small subunit of the ribosome before it binds to the m. RNA. • After the small subunit has attached to the m. RNA, it proceeds downstream (5’ to 3’) until it encounters the start codon – AUG which codes for methionine. • The region between the CAP and AUG is known as the 5’untranslated region [5’-UTR].

Initiation

The large subunit of the ribosome attaches to the small subunit, creating a second t. RNA binding site, the A site. • The A site is where the incoming aminoacyl t. RNA molecule binds (t. RNA covalently bound to its amino acid). • The P site is where the growing polypeptide chain is formed.

Elongation t. RNA charged with arginine peptide bond forms met arg new t. RNA pairs with second codon CGA UAC GCU NNN AUG G AA NN NNNNNNNNN CG second codon in A site of ribosome subunit

Elongation peptide bond forms First amino acid remains attached met NNNAUG UAC First t. RNA now detaches from ribosome arg leu new t. RNA pairs with third codon GCU GAU CGA CUA NNNNNNNNN Second codon now in P site Third codon in A site

Termination no t. RNA carries anticodon AUC met arg leu ala arg gly CCC NNNAUGCGACUAGCCAGG GGGUAG NNN m. RNA termination (stop) codon in A site

Mutations A gene mutation is a change in the nucleotide sequence of a gene.

Frame shift mutations: add or delete one or more bases from the DNA arg leu met correct reading frame A U G C G A C U A insertion met mutation A U G arg ala gly GCC AGG GGG arg gln arg ser gly CGAC G U AGC CAG GGG G G deletion met mutation A U G arg gly arg pro CGA CAG CCA GGG GG U Point mutations: involve a change in a single nucleotide and therefore a change in a specific codon

Part 7 Post Translational Modifications

a) Removal of Signal Sequence • Digestive enzymes which are secreted out of the cell are usually translated on the ribosome attached to the rough endoplasmic reticulum (RER). • The first few codons on the m. RNA encode a short signal peptide that marks the protein for export. • The signal sequence causes it to insert into the membrane surrounding the ER and push its way into the interior. • The signal sequence is removed by enzyme action and the protein is transported through the ER and Golgi apparatus to the cell.

Example of proteolytic cleavage: • Protease enzymes (that digest proteins) are produced in the pancreas to be active in the gut. If they were produced in an active form, they would digest the pancreas that produced them. Therefore they are produced as inactive preproteins and released into the gut where other enzymes activate them by cleavage.