MECHANISM OF ACTION OF NUCLEAR RECEPTORS Familias de

- Slides: 54

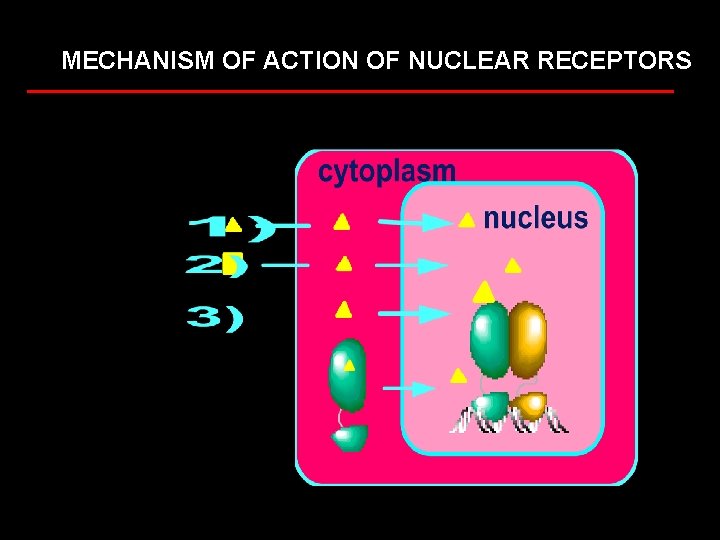
MECHANISM OF ACTION OF NUCLEAR RECEPTORS

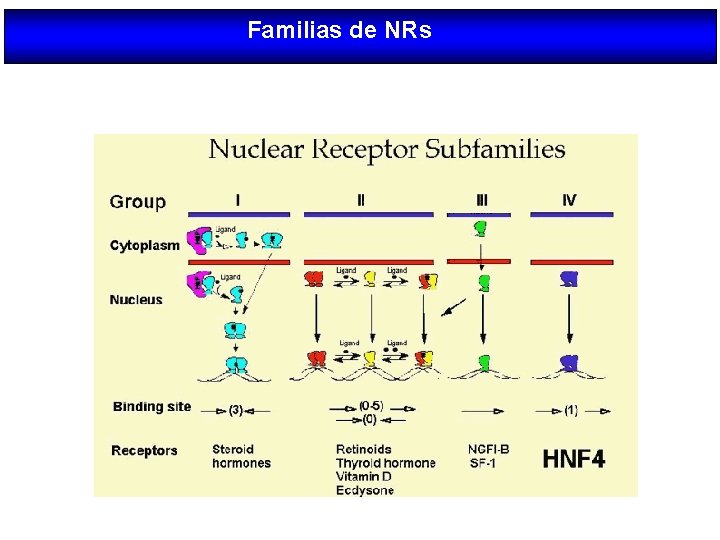
Familias de NRs

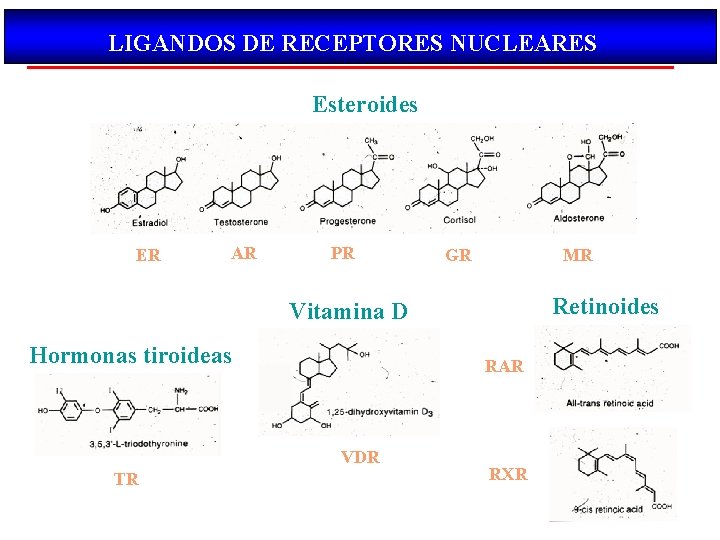
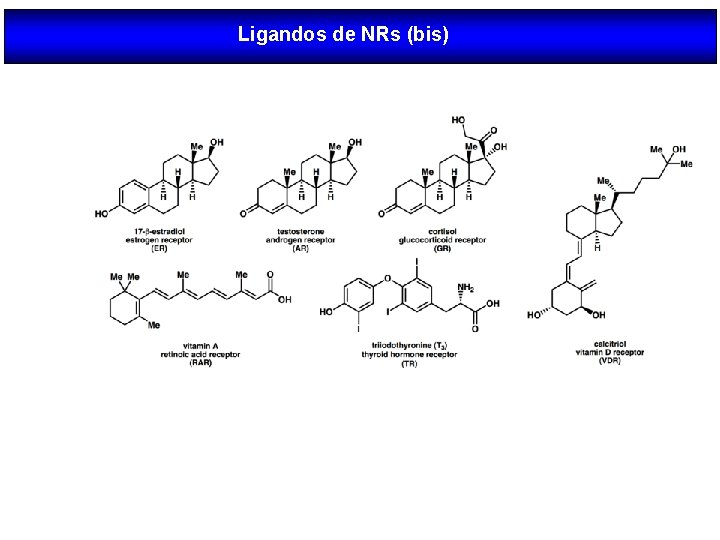
LIGANDOS DE RECEPTORES NUCLEARES Esteroides ER AR PR GR MR Retinoides Vitamina D Hormonas tiroideas RAR VDR TR RXR

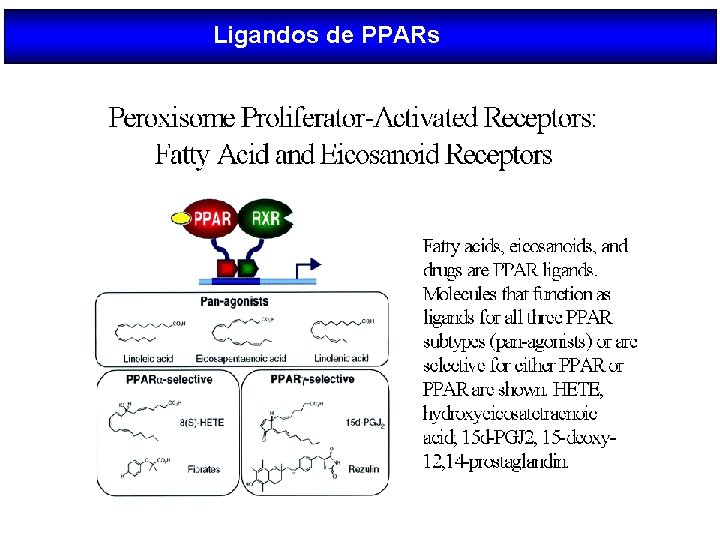
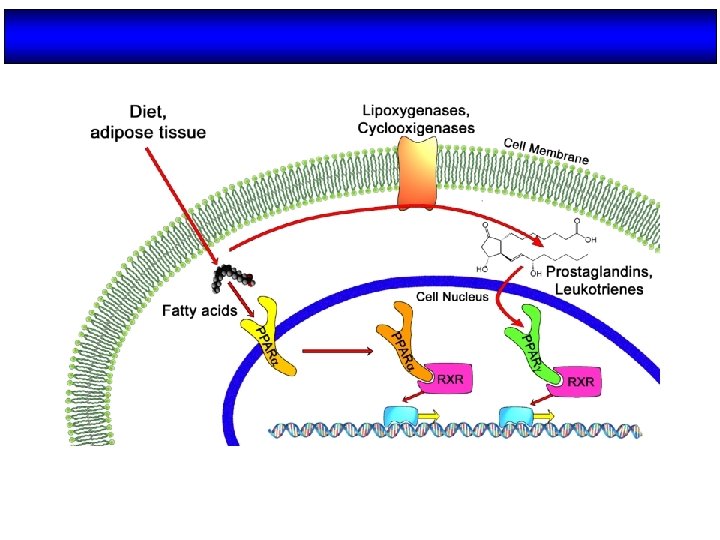
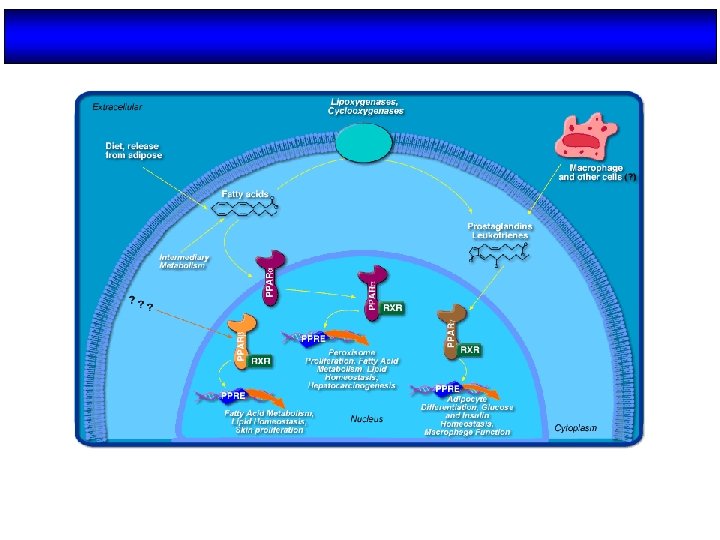
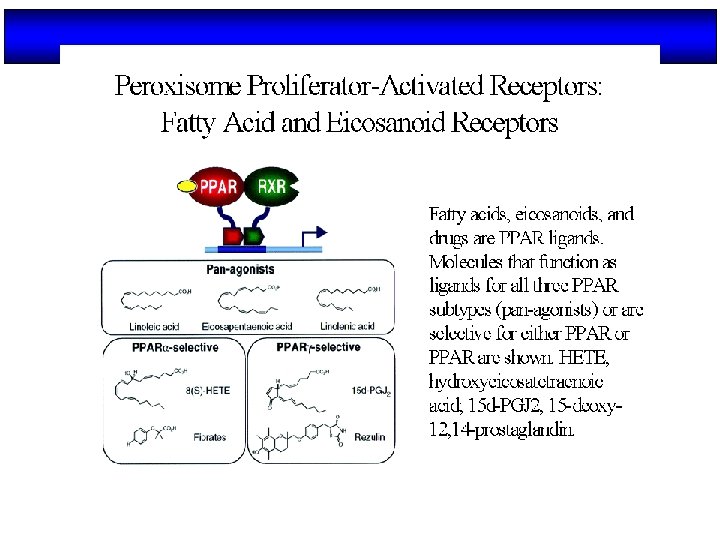
Ligandos de PPARs

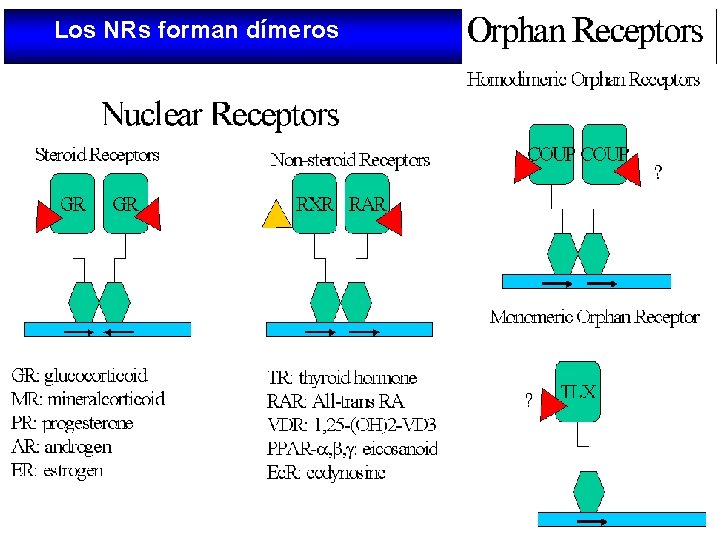
Los NRs forman dímeros

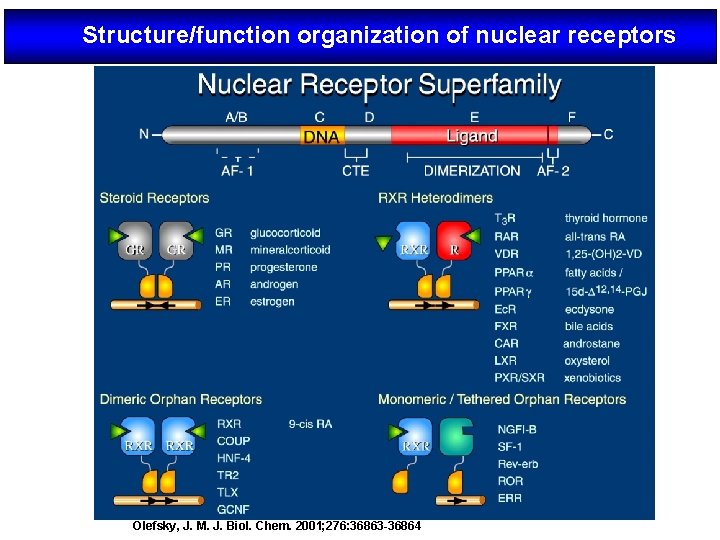
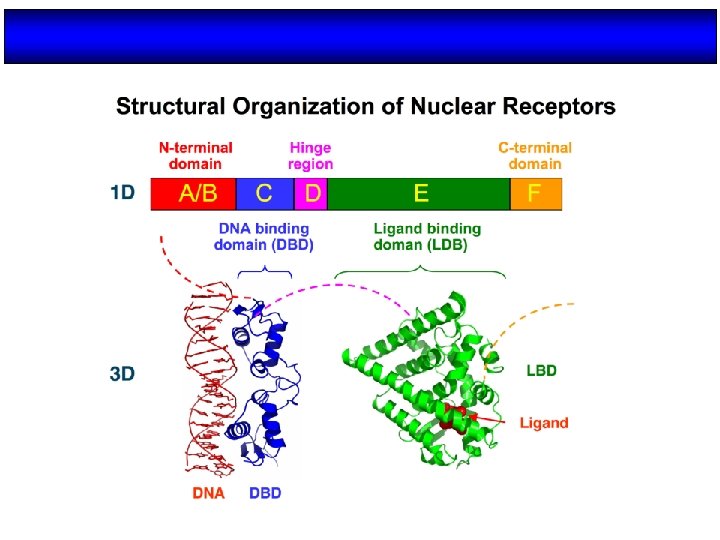
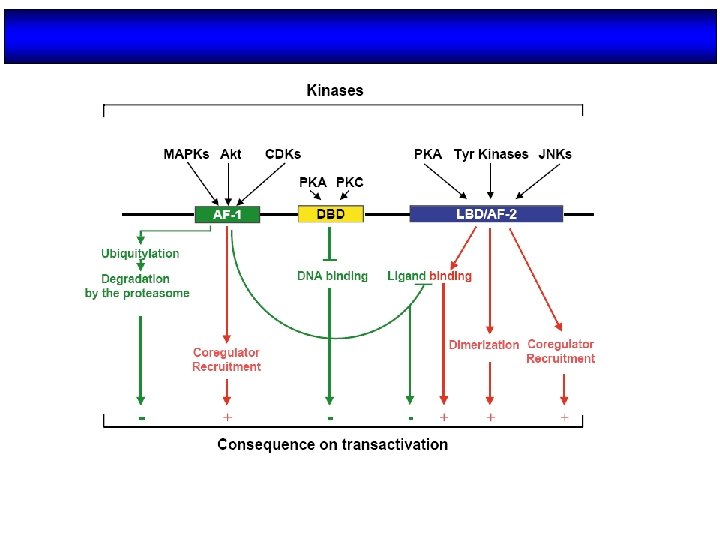
Structure/function organization of nuclear receptors Olefsky, J. M. J. Biol. Chem. 2001; 276: 36863 -36864


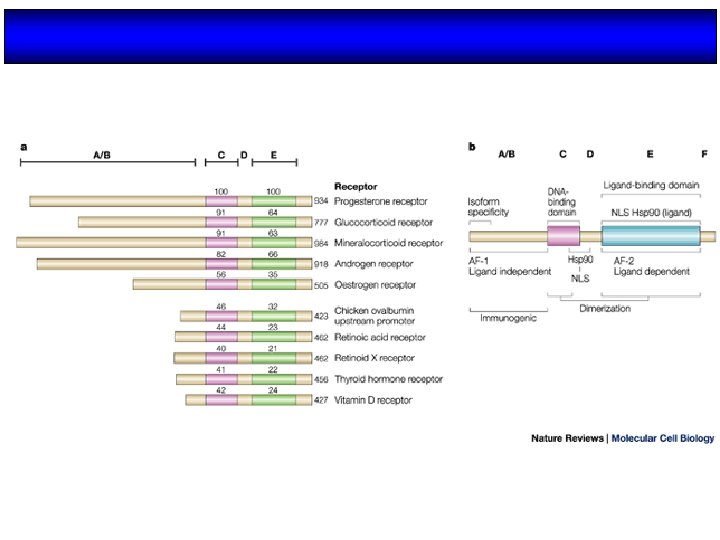
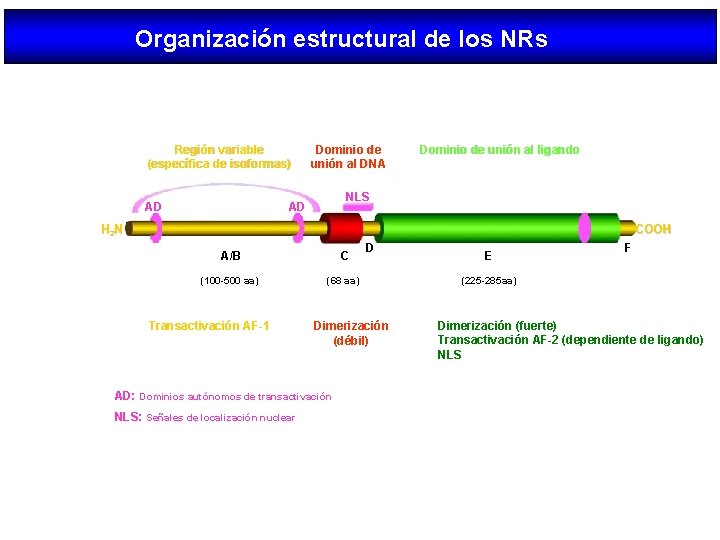
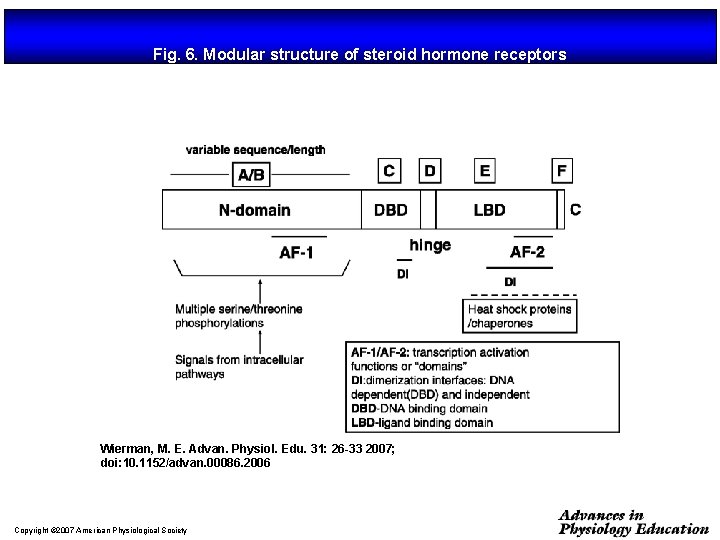
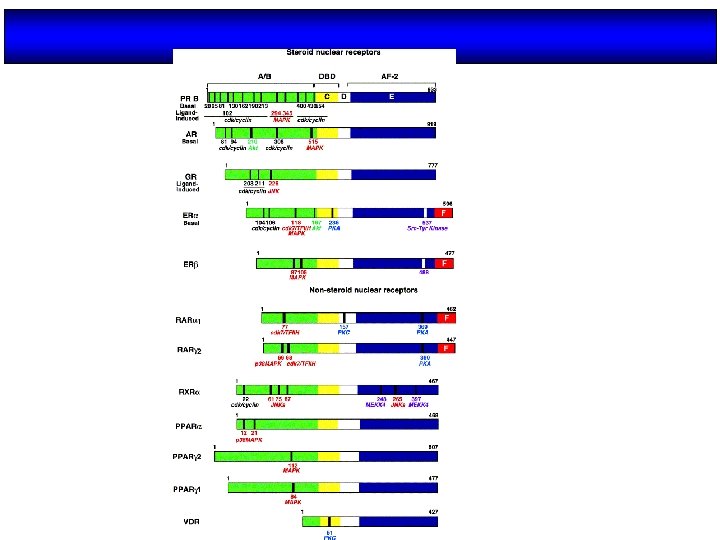
Organización estructural de los NRs Región variable (específica de isoformas) AD Dominio de unión al DNA Dominio de unión al ligando NLS AD H 2 N COOH A/B C (100 -500 aa) (68 aa) Transactivación AF-1 Dimerización (débil) AD: Dominios autónomos de transactivación NLS: Señales de localización nuclear D E F (225 -285 aa) Dimerización (fuerte) Transactivación AF-2 (dependiente de ligando) NLS


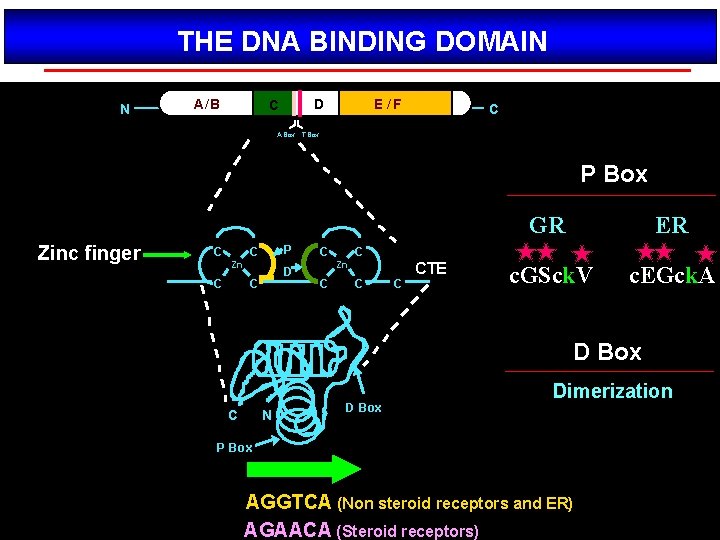
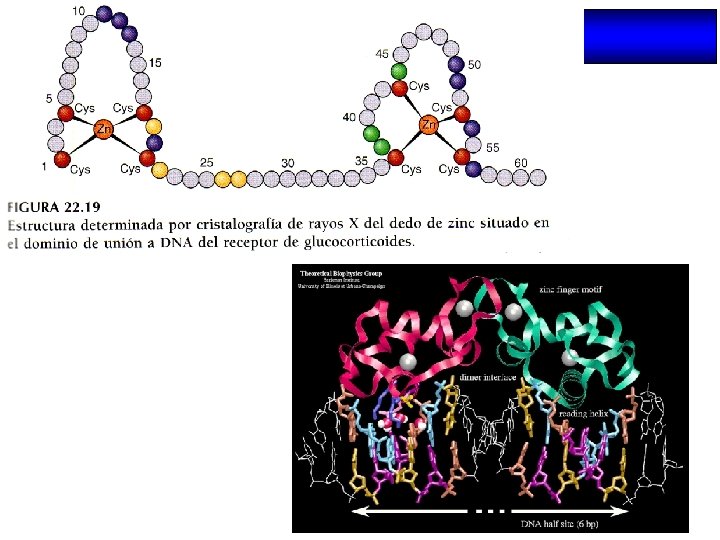
THE DNA BINDING DOMAIN N A/ B D C E /F C A Box T Box P Box Zinc finger C P C Zn C D C C ER c. GSck. V c. EGck. A C Zn C GR CTE C C D Box C N D Box Dimerization P Box AGGTCA (Non steroid receptors and ER) AGAACA (Steroid receptors)


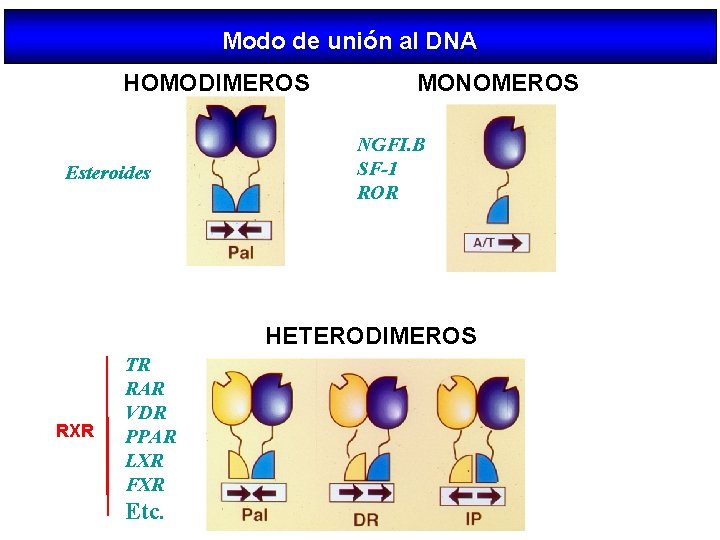
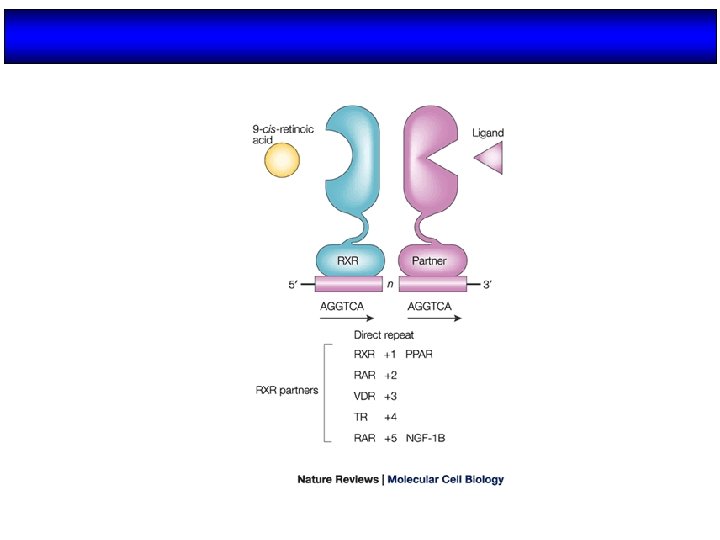
Modo de unión al DNA HOMODIMEROS Esteroides MONOMEROS NGFI. B SF-1 ROR HETERODIMEROS RXR TR RAR VDR PPAR LXR FXR Etc.

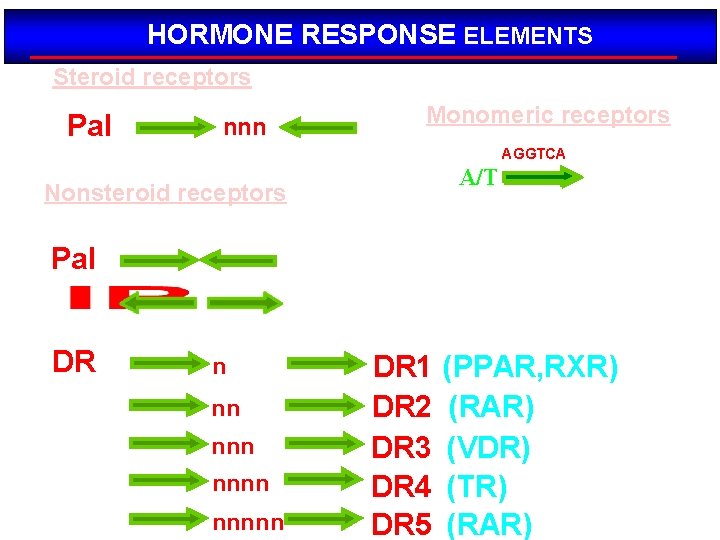
HORMONE RESPONSE ELEMENTS Steroid receptors Pal AGAACA nnn TGTTCT AGGTCA Monomeric receptors TGACCT AGGTCA A/T Nonsteroid receptors Pal DR AGGTCA n nn nnnnn DR 1 DR 2 DR 3 DR 4 DR 5 (PPAR, RXR) (RAR) (VDR) (TR) (RAR)

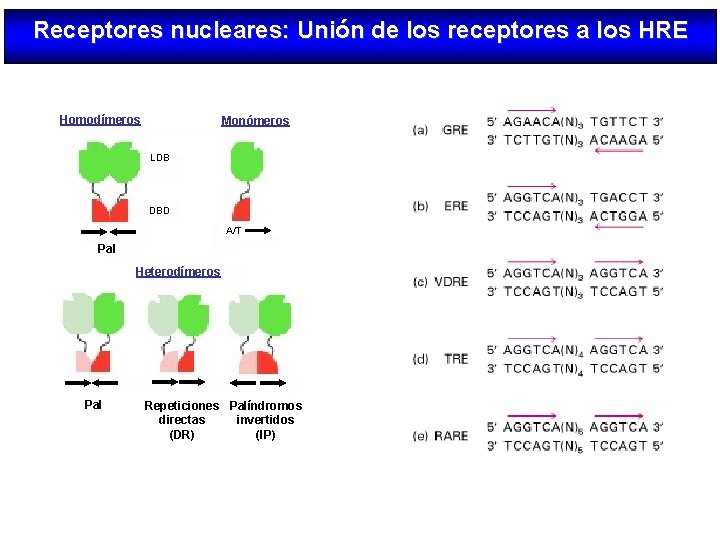
Receptores nucleares: Unión de los receptores a los HRE Homodímeros Monómeros LDB DBD A/T Pal Heterodímeros Pal Repeticiones Palíndromos directas invertidos (DR) (IP)

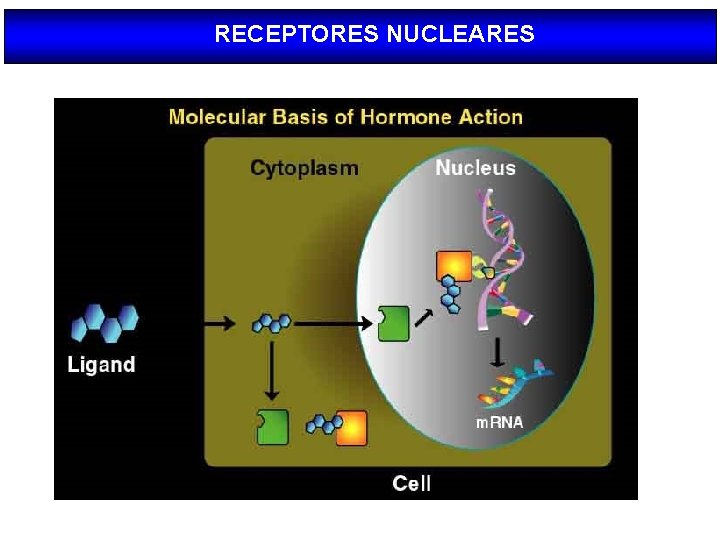
RECEPTORES NUCLEARES

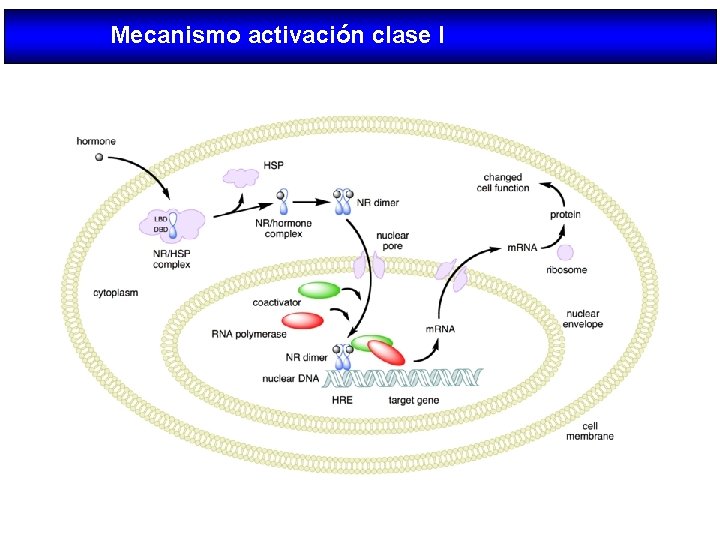
Mecanismo activación clase I

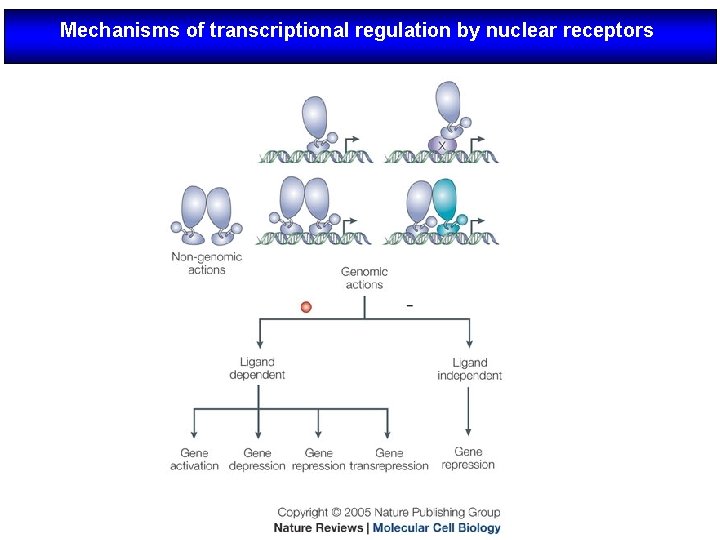
Mechanisms of transcriptional regulation by nuclear receptors

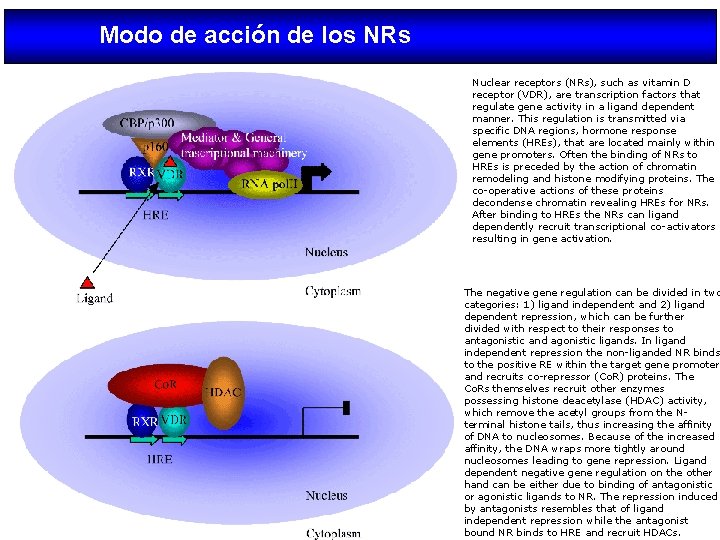
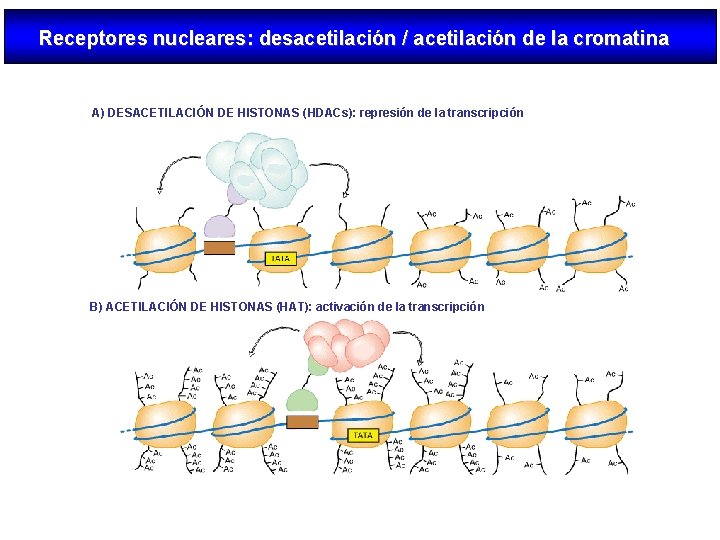
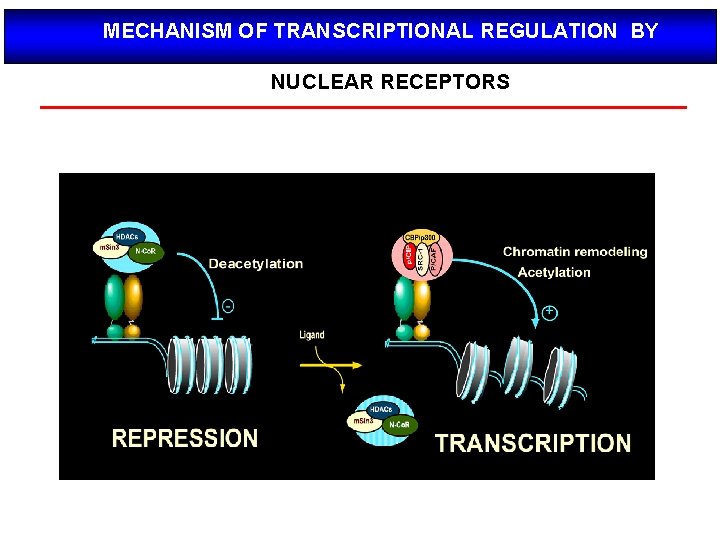
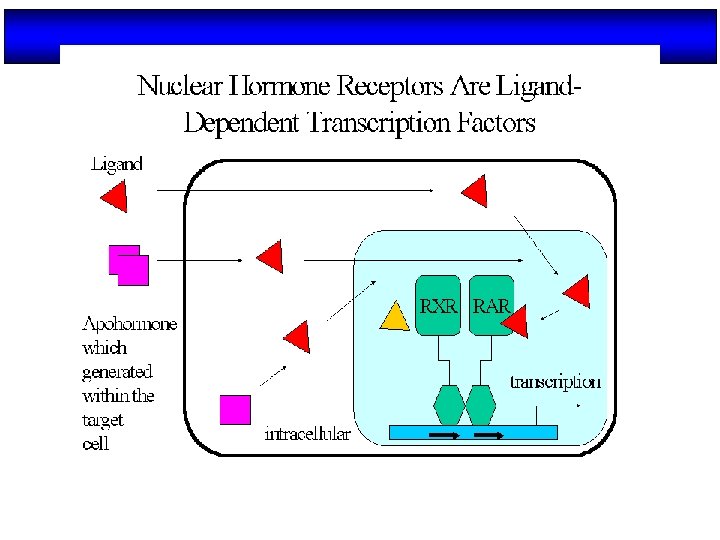
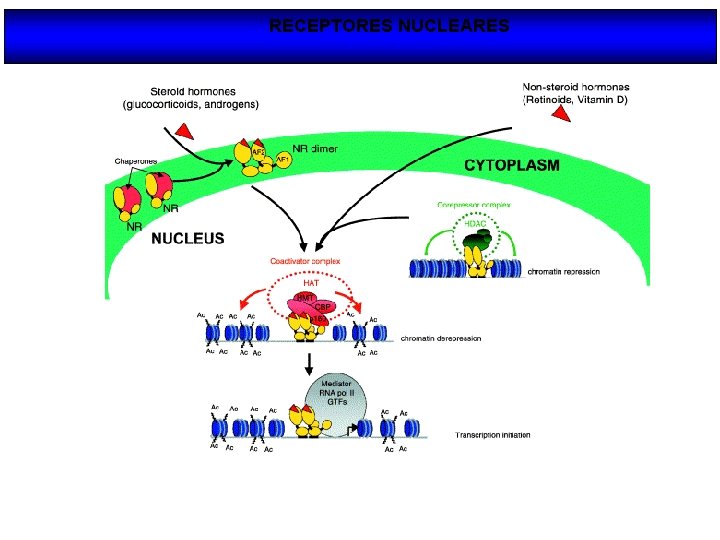
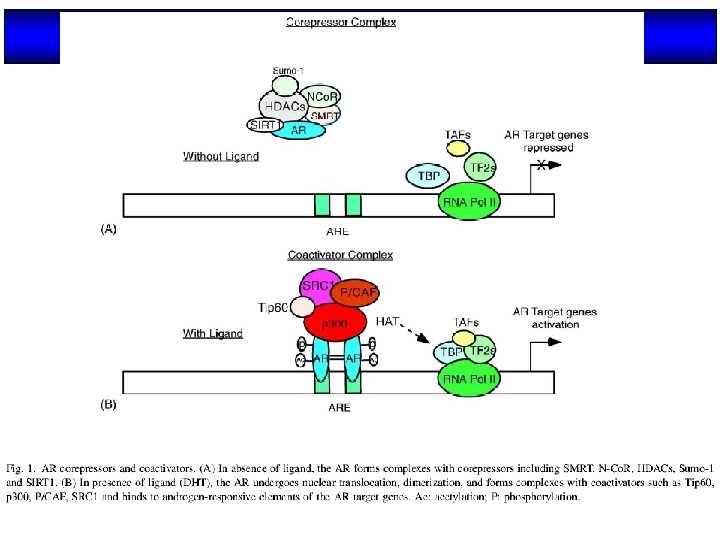
Modo de acción de los NRs Nuclear receptors (NRs), such as vitamin D receptor (VDR), are transcription factors that regulate gene activity in a ligand dependent manner. This regulation is transmitted via specific DNA regions, hormone response elements (HREs), that are located mainly within gene promoters. Often the binding of NRs to HREs is preceded by the action of chromatin remodeling and histone modifying proteins. The co-operative actions of these proteins decondense chromatin revealing HREs for NRs. After binding to HREs the NRs can ligand dependently recruit transcriptional co-activators resulting in gene activation. The negative gene regulation can be divided in two categories: 1) ligand independent and 2) ligand dependent repression, which can be further divided with respect to their responses to antagonistic and agonistic ligands. In ligand independent repression the non-liganded NR binds to the positive RE within the target gene promoter and recruits co-repressor (Co. R) proteins. The Co. Rs themselves recruit other enzymes possessing histone deacetylase (HDAC) activity, which remove the acetyl groups from the Nterminal histone tails, thus increasing the affinity of DNA to nucleosomes. Because of the increased affinity, the DNA wraps more tightly around nucleosomes leading to gene repression. Ligand dependent negative gene regulation on the other hand can be either due to binding of antagonistic or agonistic ligands to NR. The repression induced by antagonists resembles that of ligand independent repression while the antagonist bound NR binds to HRE and recruit HDACs.



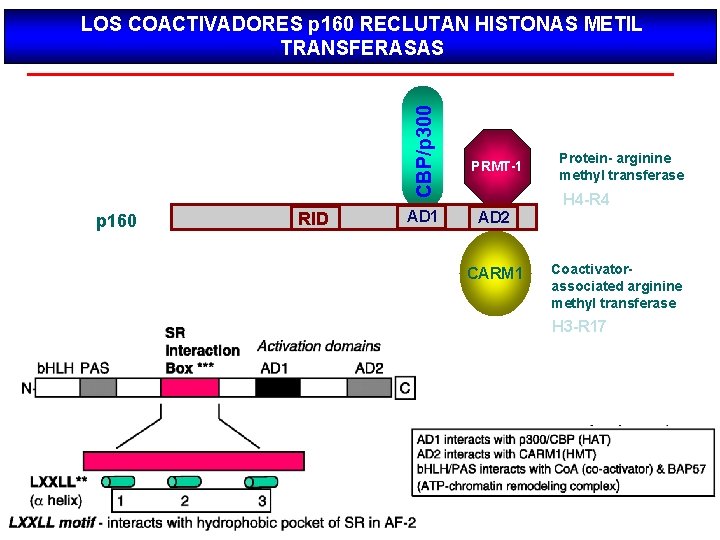
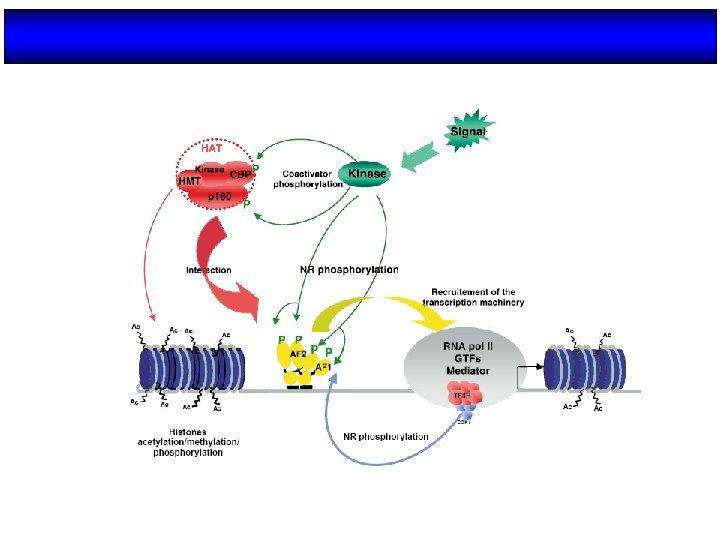
CBP/p 300 LOS COACTIVADORES p 160 RECLUTAN HISTONAS METIL TRANSFERASAS p 160 RID AD 1 PRMT-1 AD 2 CARM 1 Protein- arginine methyl transferase H 4 -R 4 Coactivatorassociated arginine methyl transferase H 3 -R 17

Receptores nucleares: desacetilación / acetilación de la cromatina A) DESACETILACIÓN DE HISTONAS (HDACs): represión de la transcripción B) ACETILACIÓN DE HISTONAS (HAT): activación de la transcripción

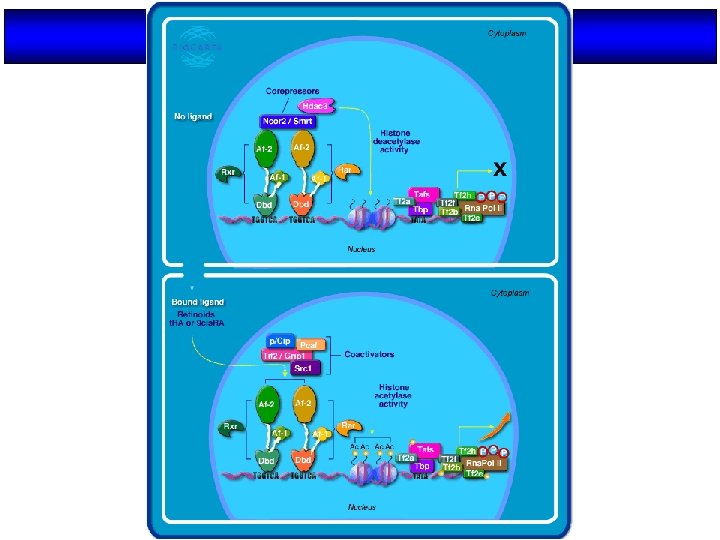
MECHANISM OF TRANSCRIPTIONAL REGULATION BY NUCLEAR RECEPTORS


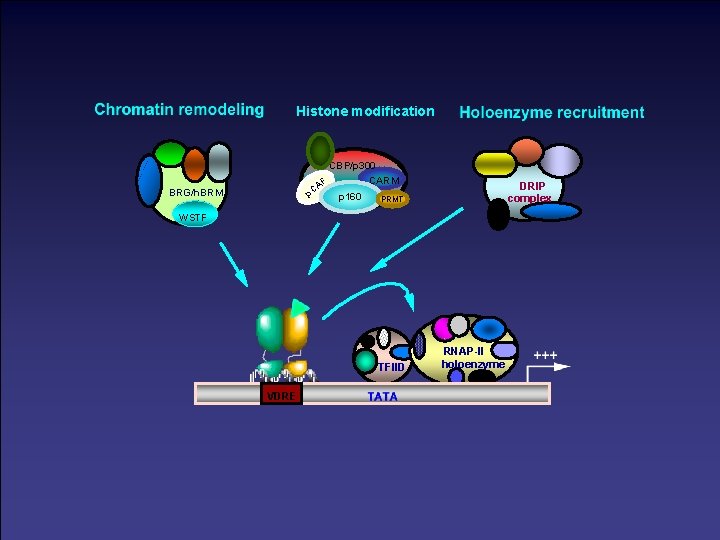
Histone modification CBP/p 300 AF p. C BRG/h. BRM CARM p 160 DRIP complex PRMT WSTF TFIID VDRE RNAP-II holoenzyme

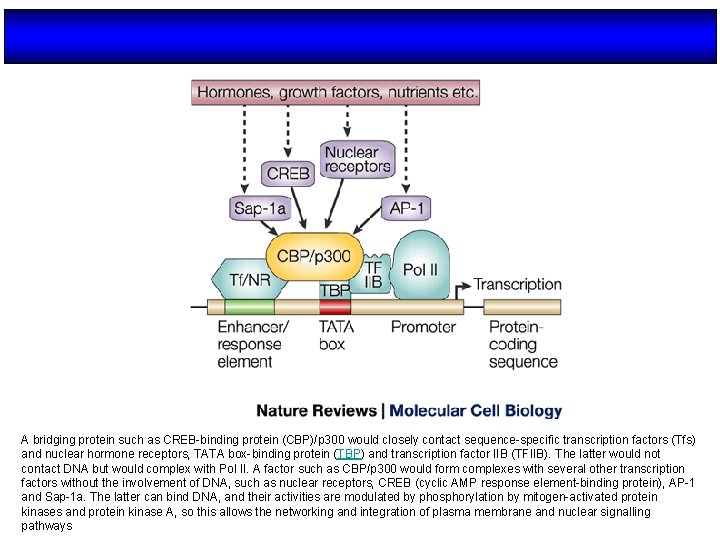
A bridging protein such as CREB-binding protein (CBP)/p 300 would closely contact sequence-specific transcription factors (Tfs) and nuclear hormone receptors, TATA box-binding protein (TBP) and transcription factor IIB (TFIIB). The latter would not contact DNA but would complex with Pol II. A factor such as CBP/p 300 would form complexes with several other transcription factors without the involvement of DNA, such as nuclear receptors, CREB (cyclic AMP response element-binding protein), AP-1 and Sap-1 a. The latter can bind DNA, and their activities are modulated by phosphorylation by mitogen-activated protein kinases and protein kinase A, so this allows the networking and integration of plasma membrane and nuclear signalling pathways

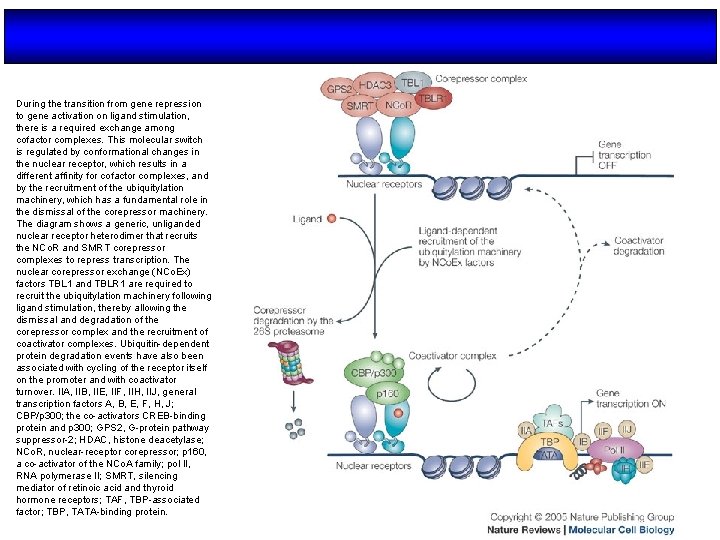
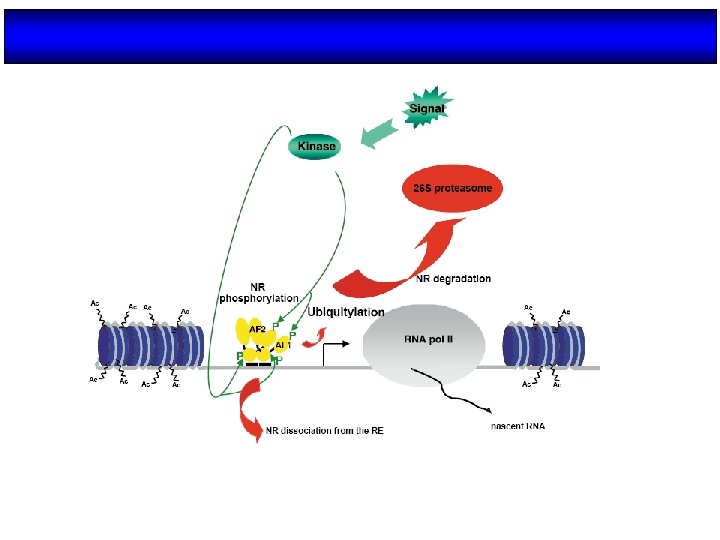
During the transition from gene repression to gene activation on ligand stimulation, there is a required exchange among cofactor complexes. This molecular switch is regulated by conformational changes in the nuclear receptor, which results in a different affinity for cofactor complexes, and by the recruitment of the ubiquitylation machinery, which has a fundamental role in the dismissal of the corepressor machinery. The diagram shows a generic, unliganded nuclear receptor heterodimer that recruits the NCo. R and SMRT corepressor complexes to repress transcription. The nuclear corepressor exchange (NCo. Ex) factors TBL 1 and TBLR 1 are required to recruit the ubiquitylation machinery following ligand stimulation, thereby allowing the dismissal and degradation of the corepressor complex and the recruitment of coactivator complexes. Ubiquitin-dependent protein degradation events have also been associated with cycling of the receptor itself on the promoter and with coactivator turnover. IIA, IIB, IIE, IIF, IIH, IIJ, general transcription factors A, B, E, F, H, J; CBP/p 300; the co-activators CREB-binding protein and p 300; GPS 2, G-protein pathway suppressor-2; HDAC, histone deacetylase; NCo. R, nuclear-receptor corepressor; p 160, a co-activator of the NCo. A family; pol II, RNA polymerase II; SMRT, silencing mediator of retinoic acid and thyroid hormone receptors; TAF, TBP-associated factor; TBP, TATA-binding protein.

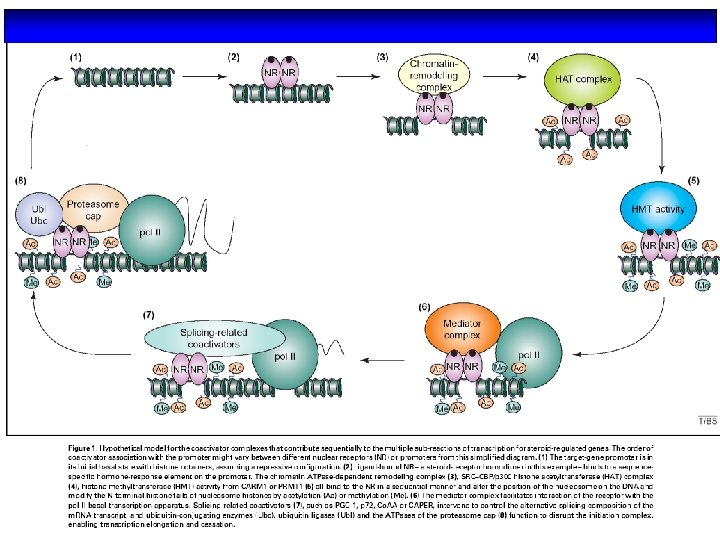
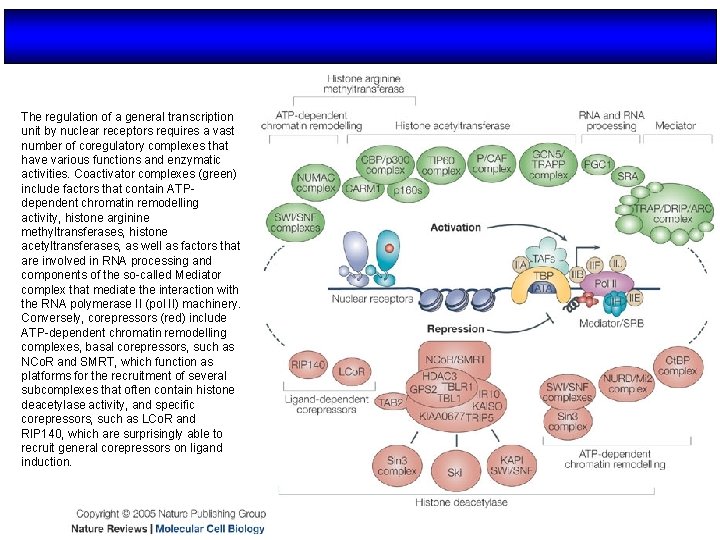
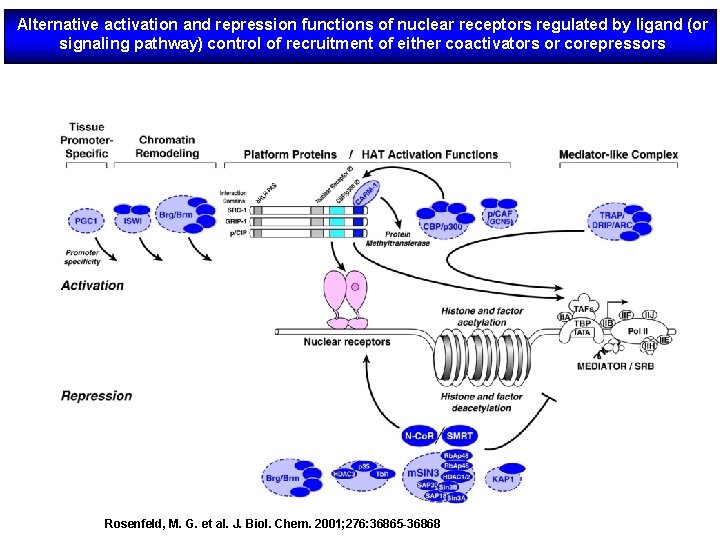
The regulation of a general transcription unit by nuclear receptors requires a vast number of coregulatory complexes that have various functions and enzymatic activities. Coactivator complexes (green) include factors that contain ATPdependent chromatin remodelling activity, histone arginine methyltransferases, histone acetyltransferases, as well as factors that are involved in RNA processing and components of the so-called Mediator complex that mediate the interaction with the RNA polymerase II (pol II) machinery. Conversely, corepressors (red) include ATP-dependent chromatin remodelling complexes, basal corepressors, such as NCo. R and SMRT, which function as platforms for the recruitment of several subcomplexes that often contain histone deacetylase activity, and specific corepressors, such as LCo. R and RIP 140, which are surprisingly able to recruit general corepressors on ligand induction.

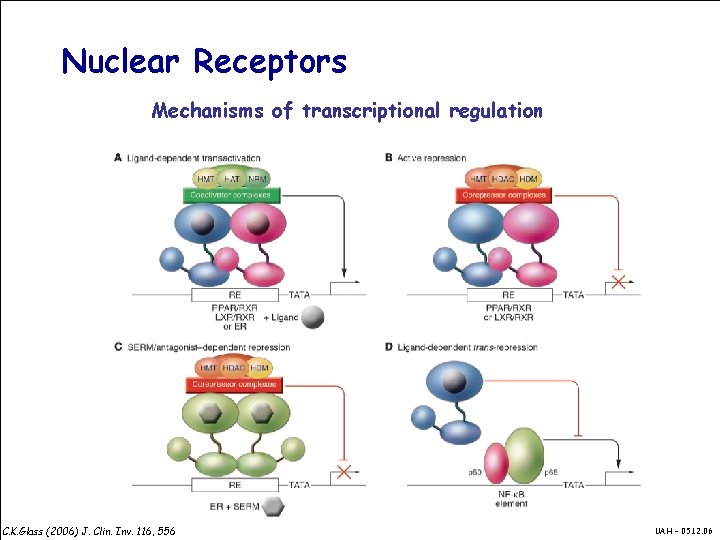
Nuclear Receptors Mechanisms of transcriptional regulation C. K. Glass (2006) J. Clin. Inv. 116, 556 UAH – 05. 12. 06


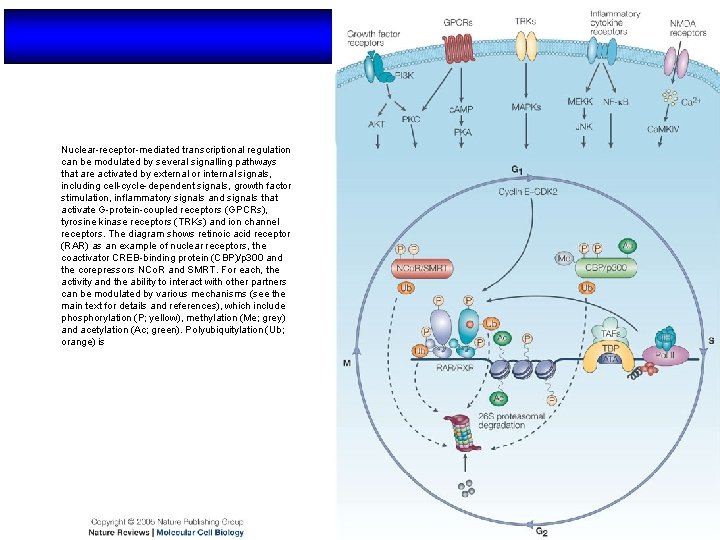
Nuclear-receptor-mediated transcriptional regulation can be modulated by several signalling pathways that are activated by external or internal signals, including cell-cycle-dependent signals, growth factor stimulation, inflammatory signals and signals that activate G-protein-coupled receptors (GPCRs), tyrosine kinase receptors (TRKs) and ion channel receptors. The diagram shows retinoic acid receptor (RAR) as an example of nuclear receptors, the coactivator CREB-binding protein (CBP)/p 300 and the corepressors NCo. R and SMRT. For each, the activity and the ability to interact with other partners can be modulated by various mechanisms (see the main text for details and references), which include phosphorylation (P; yellow), methylation (Me; grey) and acetylation (Ac; green). Polyubiquitylation (Ub; orange) is



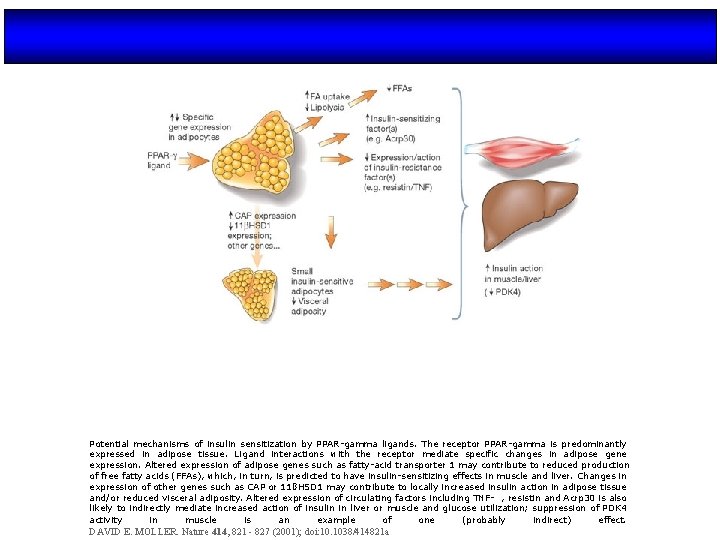
Potential mechanisms of insulin sensitization by PPAR-gamma ligands. The receptor PPAR-gamma is predominantly expressed in adipose tissue. Ligand interactions with the receptor mediate specific changes in adipose gene expression. Altered expression of adipose genes such as fatty-acid transporter 1 may contribute to reduced production of free fatty acids (FFAs), which, in turn, is predicted to have insulin-sensitizing effects in muscle and liver. Changes in expression of other genes such as CAP or 11ßHSD 1 may contribute to locally increased insulin action in adipose tissue and/or reduced visceral adiposity. Altered expression of circulating factors including TNF- , resistin and Acrp 30 is also likely to indirectly mediate increased action of insulin in liver or muscle and glucose utilization; suppression of PDK 4 activity in muscle is an example of one (probably indirect) effect. DAVID E. MOLLER. Nature 414, 821 - 827 (2001); doi: 10. 1038/414821 a

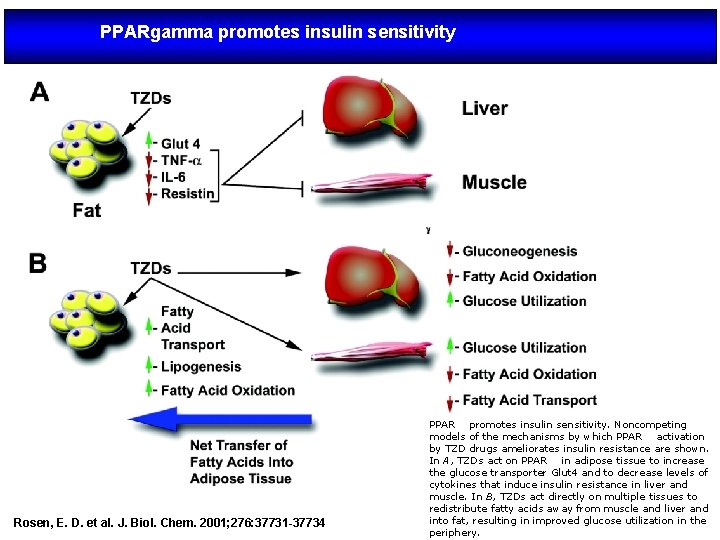
PPARgamma promotes insulin sensitivity Rosen, E. D. et al. J. Biol. Chem. 2001; 276: 37731 -37734 PPAR promotes insulin sensitivity. Noncompeting models of the mechanisms by which PPAR activation by TZD drugs ameliorates insulin resistance are shown. In A, TZDs act on PPAR in adipose tissue to increase the glucose transporter Glut 4 and to decrease levels of cytokines that induce insulin resistance in liver and muscle. In B, TZDs act directly on multiple tissues to redistribute fatty acids away from muscle and liver and into fat, resulting in improved glucose utilization in the periphery.




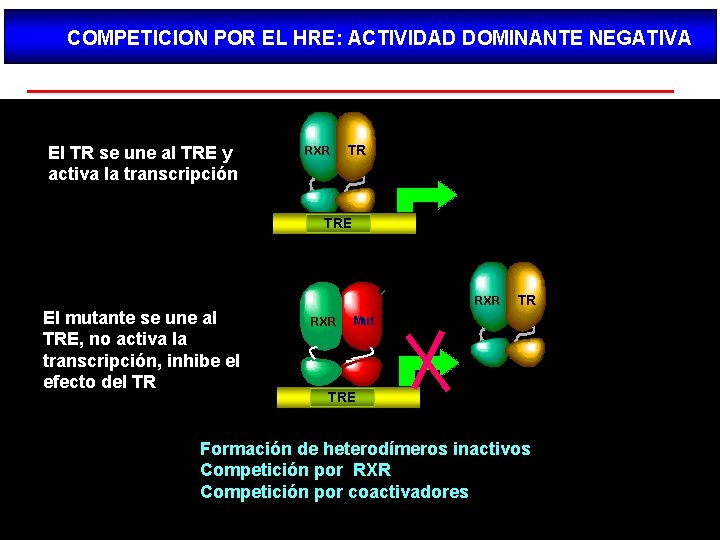
COMPETICION POR EL HRE: ACTIVIDAD DOMINANTE NEGATIVA El TR se une al TRE y activa la transcripción RXR TR TRE El mutante se une al TRE, no activa la transcripción, inhibe el efecto del TR RXR TR Mut TRE Formación de heterodímeros inactivos Competición por RXR Competición por coactivadores

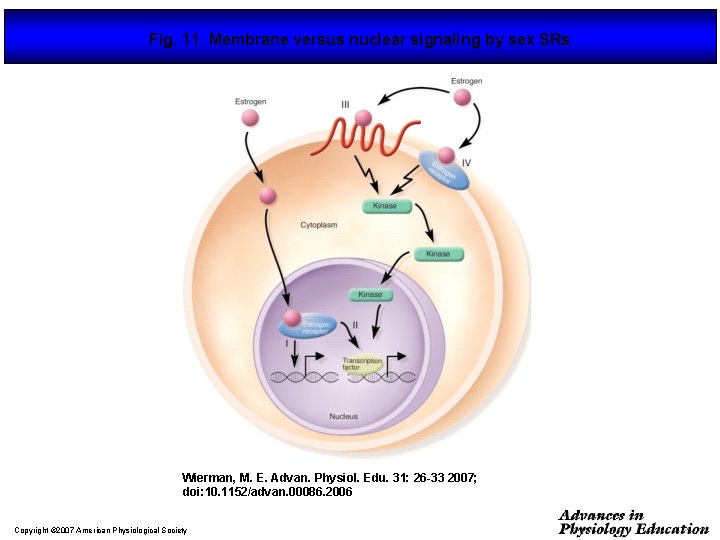
Fig. 11. Membrane versus nuclear signaling by sex SRs Wierman, M. E. Advan. Physiol. Edu. 31: 26 -33 2007; doi: 10. 1152/advan. 00086. 2006 Copyright © 2007 American Physiological Society





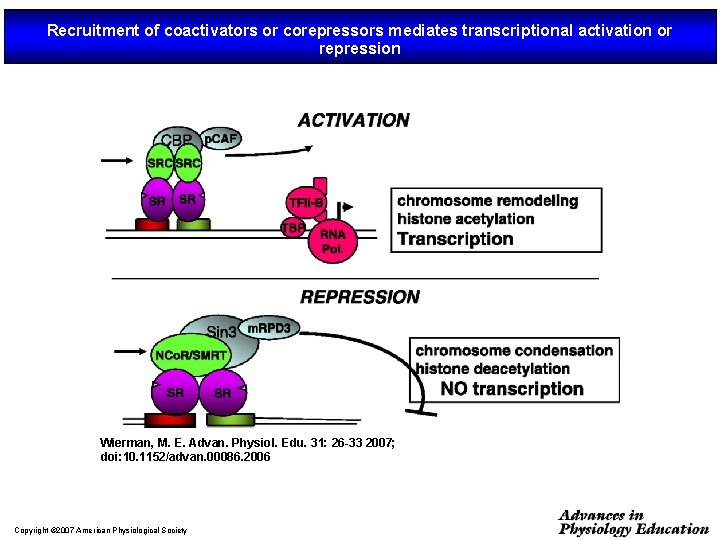
Recruitment of coactivators or corepressors mediates transcriptional activation or repression Wierman, M. E. Advan. Physiol. Edu. 31: 26 -33 2007; doi: 10. 1152/advan. 00086. 2006 Copyright © 2007 American Physiological Society

Fig. 6. Modular structure of steroid hormone receptors Wierman, M. E. Advan. Physiol. Edu. 31: 26 -33 2007; doi: 10. 1152/advan. 00086. 2006 Copyright © 2007 American Physiological Society

Alternative activation and repression functions of nuclear receptors regulated by ligand (or signaling pathway) control of recruitment of either coactivators or corepressors Rosenfeld, M. G. et al. J. Biol. Chem. 2001; 276: 36865 -36868

RECEPTORES NUCLEARES


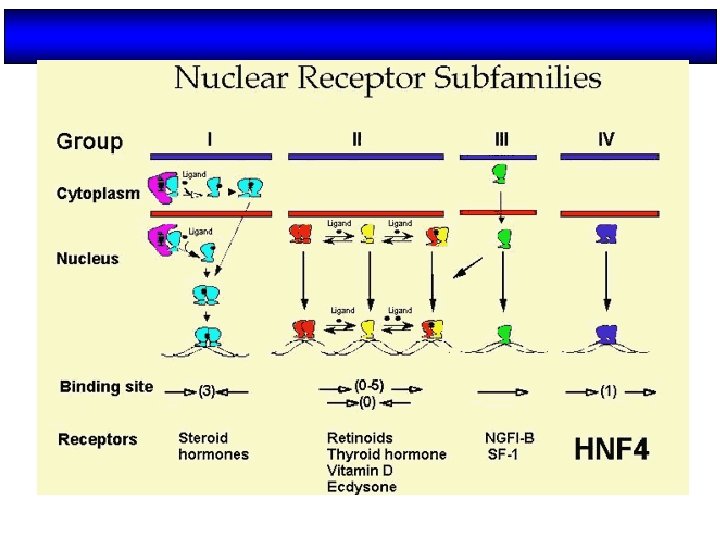
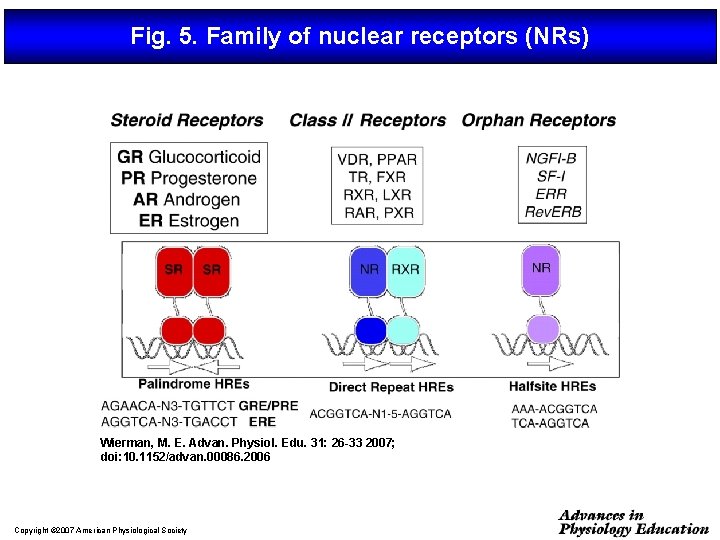
Fig. 5. Family of nuclear receptors (NRs) Wierman, M. E. Advan. Physiol. Edu. 31: 26 -33 2007; doi: 10. 1152/advan. 00086. 2006 Copyright © 2007 American Physiological Society

Ligandos de NRs (bis)


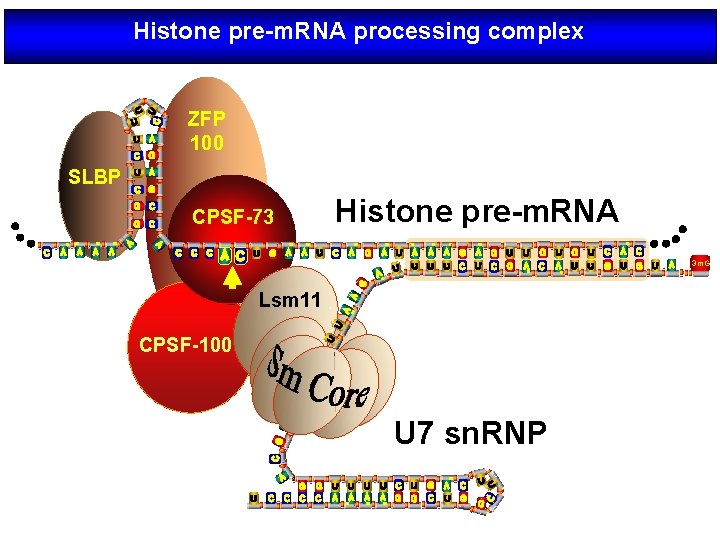
Histone pre-m. RNA processing complex ZFP 100 SLBP CPSF-73 Histone pre-m. RNA 3 m. G Lsm 11 CPSF-100 U 7 sn. RNP

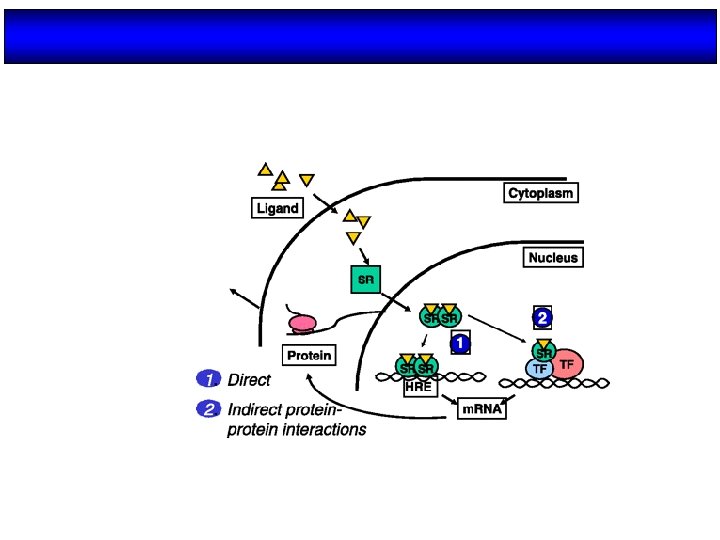
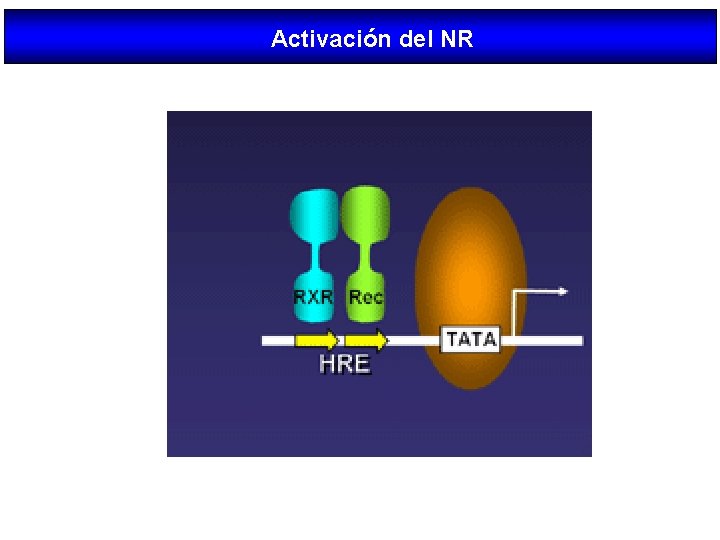
Activación del NR