Measuring Evolution Evidence for evolution Estimate natural selection





- Slides: 24

Measuring Evolution • Evidence for evolution – Estimate natural selection in the wild – Trace fossils – Infer phylogeny from behavior – Infer behavioral evolution from phylogeny • The comparative method – Test evolutionary hypotheses by correlating behavior with ecological factors across • Species • Phylogenetically independent contrasts

Selection and Galapagos finches 14 species evolved in about 500, 000 years

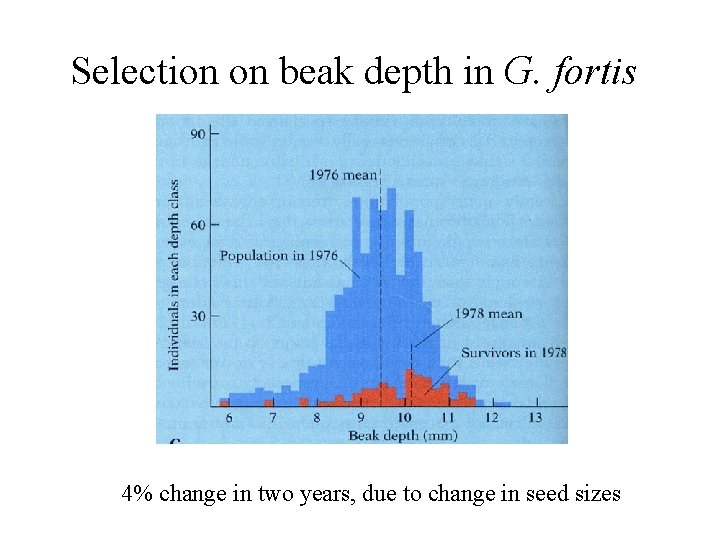
Selection on beak depth in G. fortis 4% change in two years, due to change in seed sizes

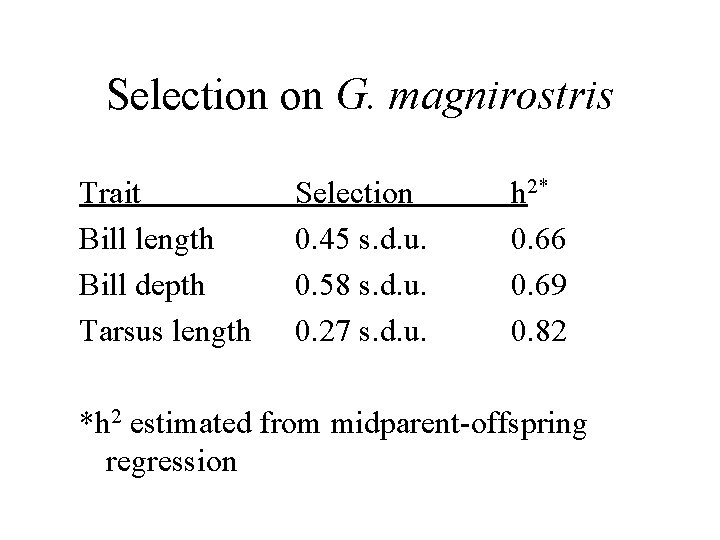
Selection on G. magnirostris Trait Bill length Bill depth Tarsus length Selection 0. 45 s. d. u. 0. 58 s. d. u. 0. 27 s. d. u. h 2* 0. 66 0. 69 0. 82 *h 2 estimated from midparent-offspring regression

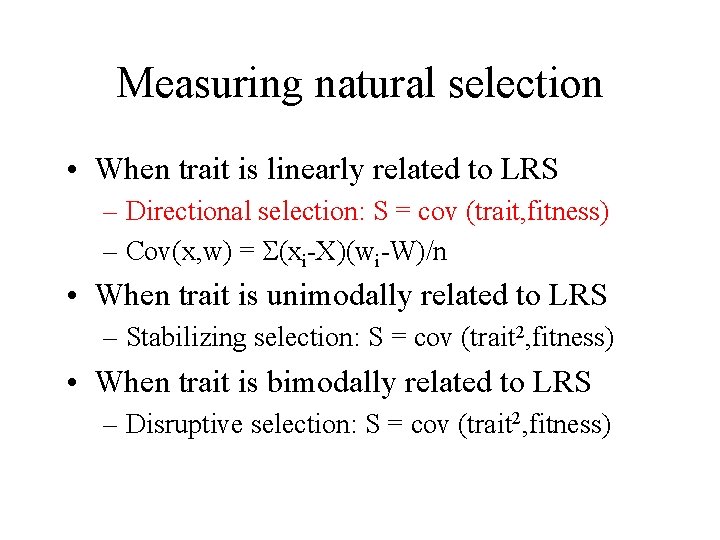
Measuring natural selection • When trait is linearly related to LRS – Directional selection: S = cov (trait, fitness) – Cov(x, w) = S(xi-X)(wi-W)/n • When trait is unimodally related to LRS – Stabilizing selection: S = cov (trait 2, fitness) • When trait is bimodally related to LRS – Disruptive selection: S = cov (trait 2, fitness)

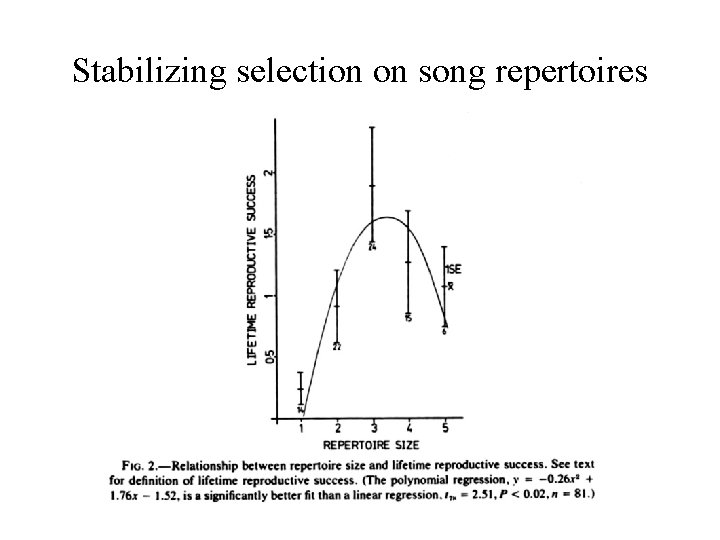
Stabilizing selection on song repertoires

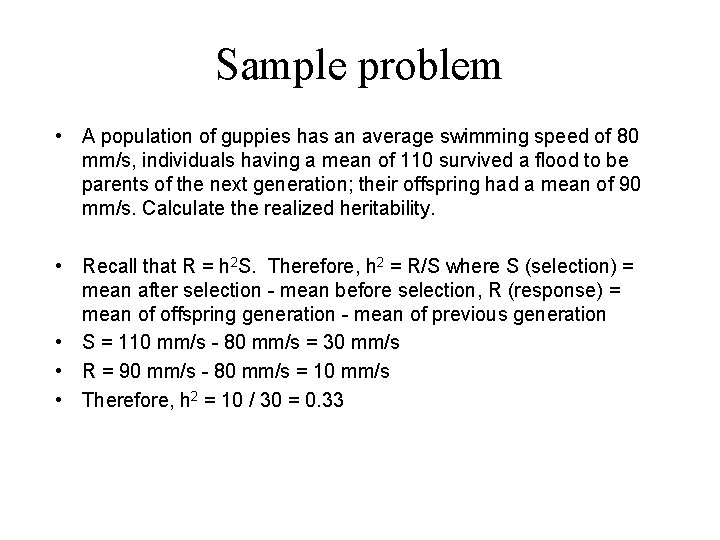
Sample problem • A population of guppies has an average swimming speed of 80 mm/s, individuals having a mean of 110 survived a flood to be parents of the next generation; their offspring had a mean of 90 mm/s. Calculate the realized heritability. • Recall that R = h 2 S. Therefore, h 2 = R/S where S (selection) = mean after selection - mean before selection, R (response) = mean of offspring generation - mean of previous generation • S = 110 mm/s - 80 mm/s = 30 mm/s • R = 90 mm/s - 80 mm/s = 10 mm/s • Therefore, h 2 = 10 / 30 = 0. 33

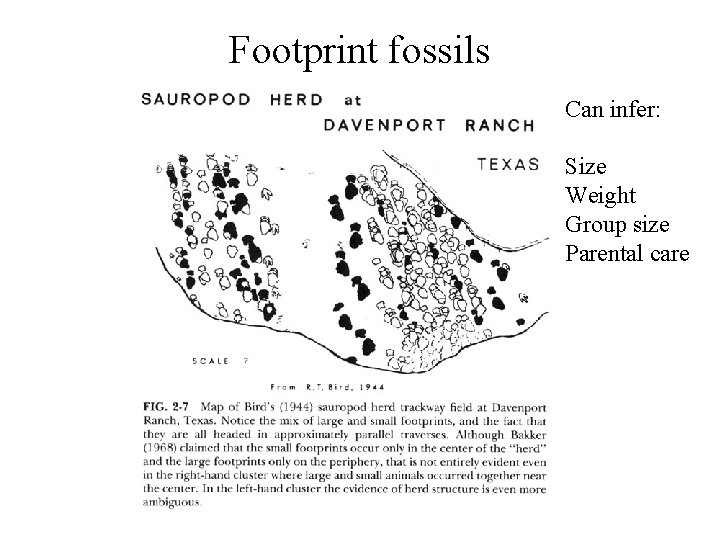
Footprint fossils Can infer: Size Weight Group size Parental care

Homo erectus footprints 3. 8 MYA Laetoli, Tanzania

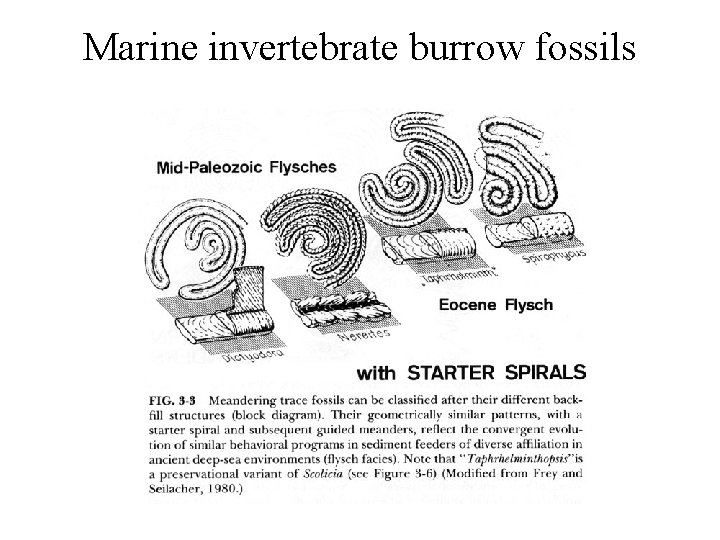
Marine invertebrate burrow fossils

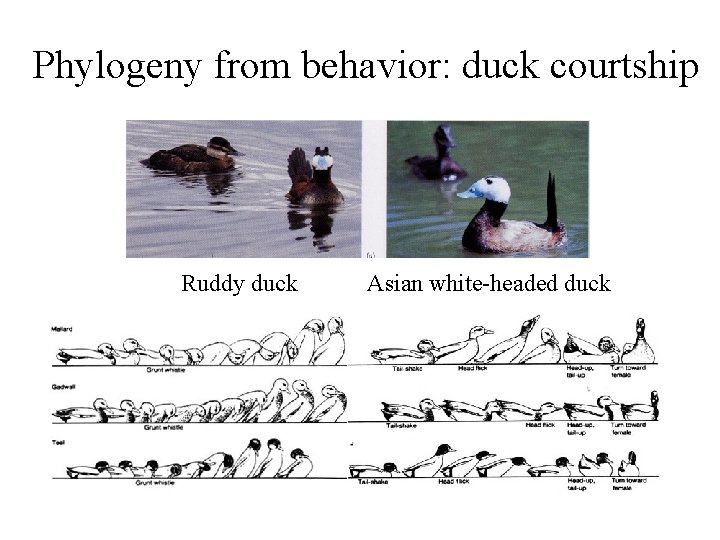
Phylogeny from behavior: duck courtship Ruddy duck Asian white-headed duck

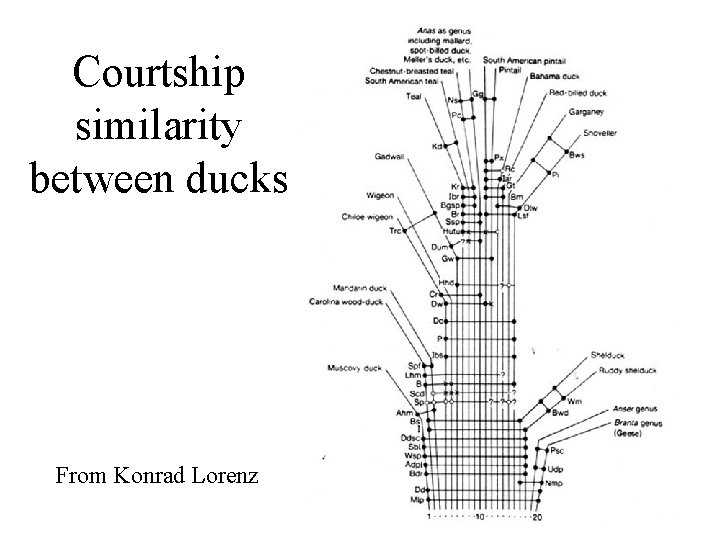
Courtship similarity between ducks From Konrad Lorenz

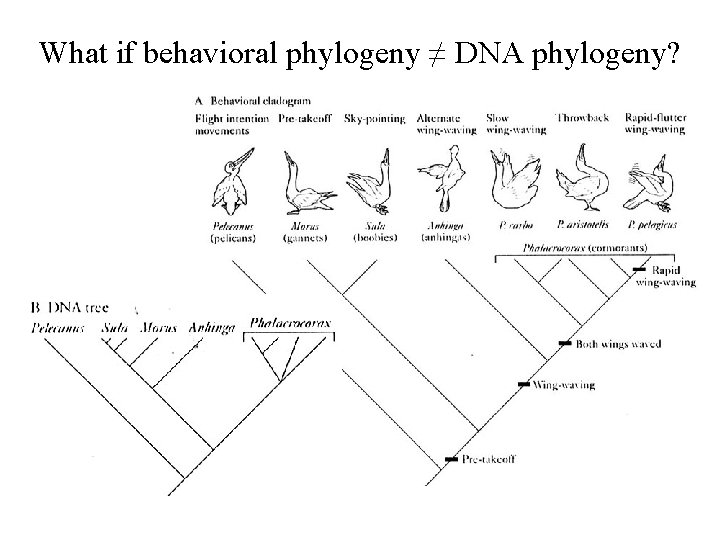
What if behavioral phylogeny ≠ DNA phylogeny?

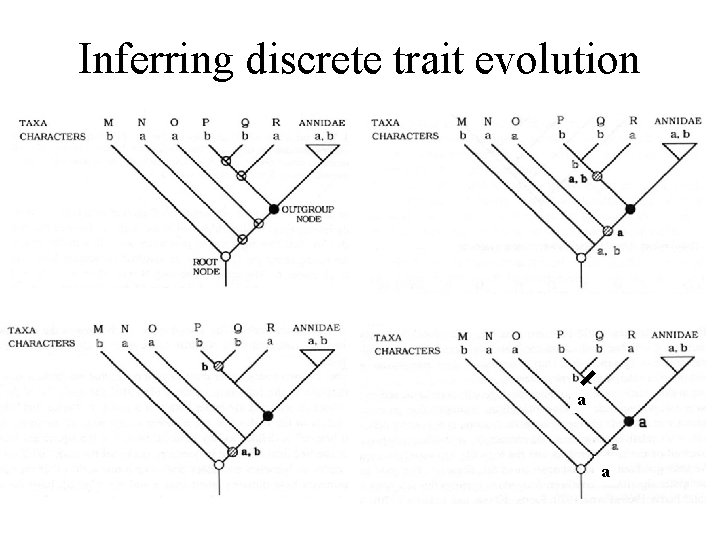
Infer trait evolution • Derive phylogenetic tree from independent data • Assign trait values to ancestral nodes by minimizing the number of possible changes, i. e. use parsimony • Use trait value at next lower node to decide ambiguous nodes • Note: need outgroup to infer ancestral trait value

Inferring discrete trait evolution a a

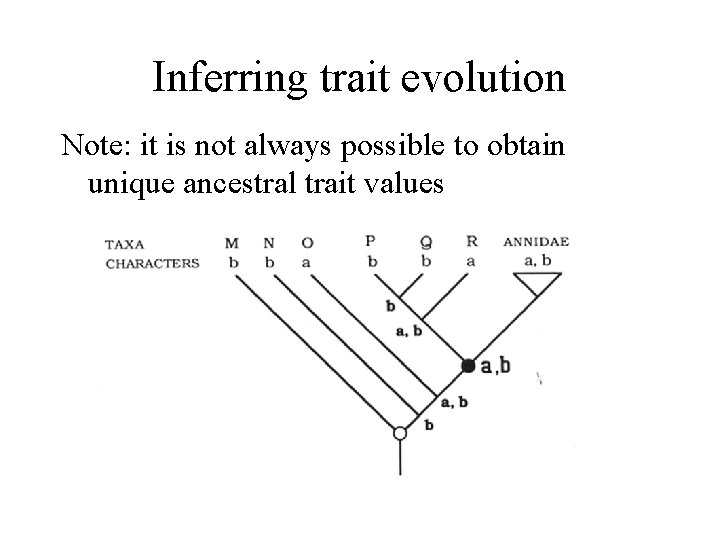
Inferring trait evolution Note: it is not always possible to obtain unique ancestral trait values

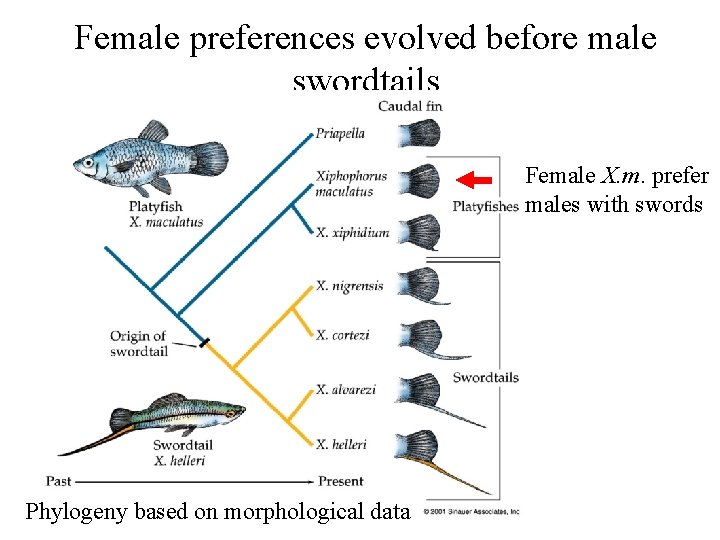
Phylogenies can be used to test adaptationist hypotheses Sexual selection example – Sensory exploitation hyothesis • Female preference evolves prior to male trait – Coevolution hypothesis • Female preference evolves with male trait

Female preferences evolved before male swordtails Female X. m. prefer males with swords Phylogeny based on morphological data

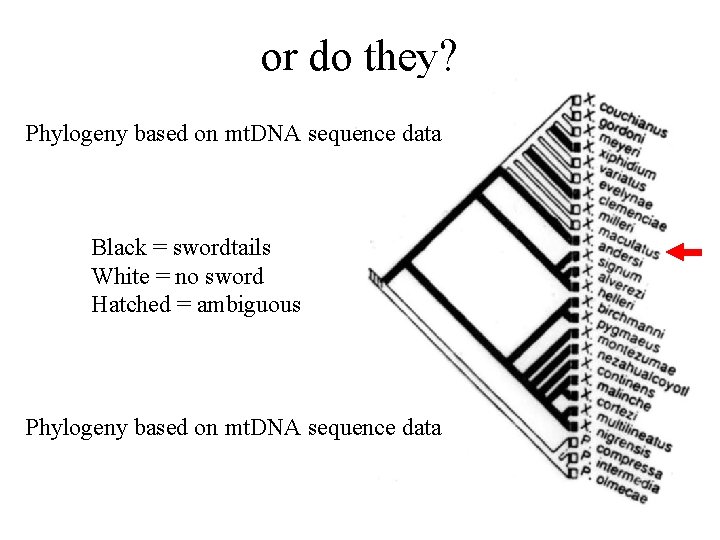
or do they? Phylogeny based on mt. DNA sequence data Black = swordtails White = no sword Hatched = ambiguous Phylogeny based on mt. DNA sequence data

The comparative method and continuous characters • Predict that some trait correlates with a particular ecological or social variable due to selection • Remove effects of body size, if necessary • Look for correlation between traits across species • Assumes species are independent, unlikely!

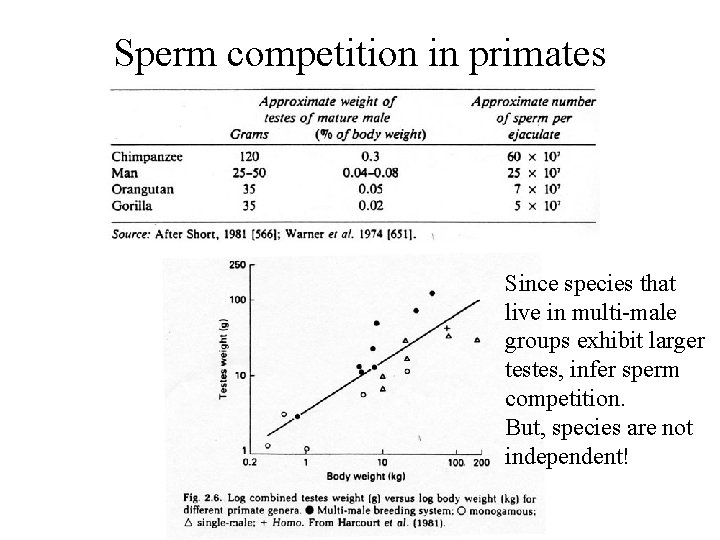
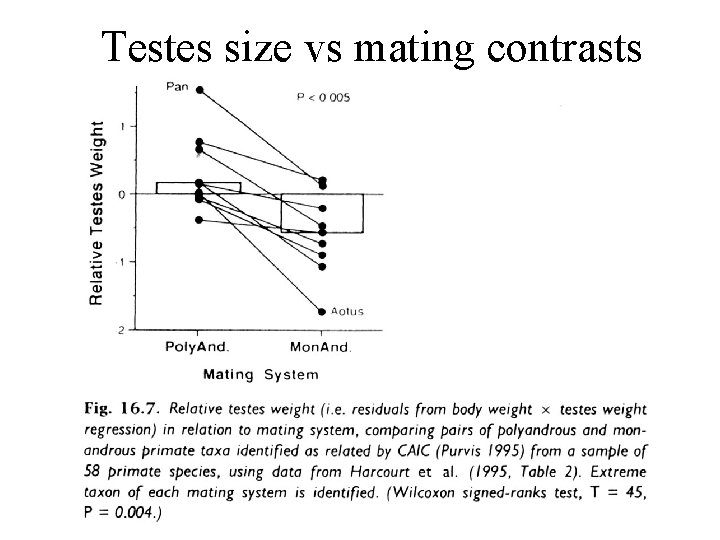
Sperm competition in primates Since species that live in multi-male groups exhibit larger testes, infer sperm competition. But, species are not independent!

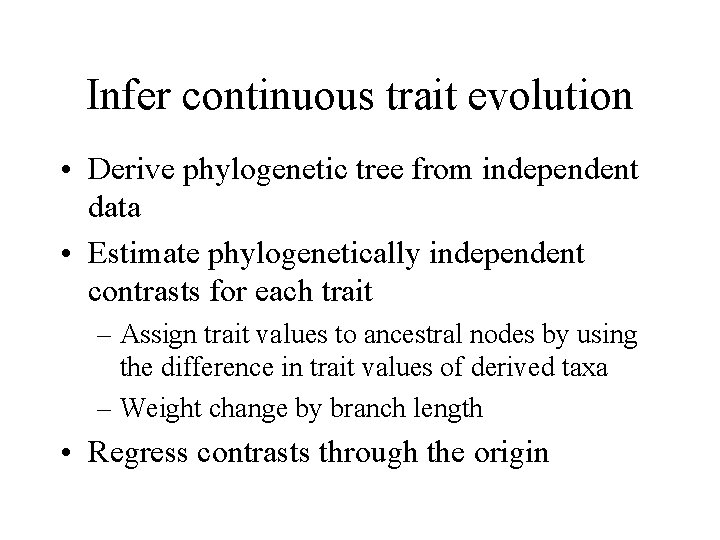
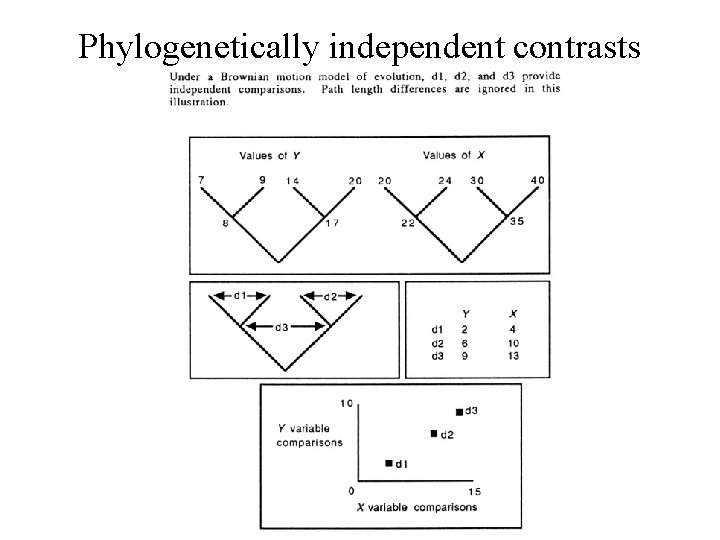
Infer continuous trait evolution • Derive phylogenetic tree from independent data • Estimate phylogenetically independent contrasts for each trait – Assign trait values to ancestral nodes by using the difference in trait values of derived taxa – Weight change by branch length • Regress contrasts through the origin

Phylogenetically independent contrasts

Testes size vs mating contrasts