MCB 3637 Lecture 16 Nov 1518 Read alignment

MCB 3637 Lecture #16 Nov 15/18 Read alignment and SNP calling

Reference mapping • Align all of the reads from your strain to a reference genome that has already been sequenced and assembled • Assumes that the genome that you sequenced is closely related to the reference genome • Computationally quite fast,

Basic workflow 1. Download reads and the reference genome 2. Align reads to the reference genome 3. Are there any differences between the sequenced genome and the reference genome? • Many steps in the pipeline, collate together as a script • Efficient & reproducible

Read mapping tools • Many different flavors • bwa (we will use today) • Some debate about which is best, trade-offs between sensitivity (ability to map everything) and specificity (are mappings correct) • Also speed and memory considerations

Mapping using bwa • Load bwa module: module load bwa • Create an index of the reference genome nucleotide fasta for the alignment software to use for read mapping • syntax: bwa index [ref. fasta file] • e. g. : bwa index E_coli. fasta • note: use ". fasta" file ending for a later step • Creates 5 output files: [ref. fasta]. amb, . ann, . bwt, . pac, . sa • "Index": special computer data structure that allows fast searching; software-specific

Mapping using bwa • bwa mem does the actual mapping step • syntax: bwa mem -R '@RGt. ID: [name 1]t. SM: [name 2]t. LB: libra ry 1' [ref. fasta file] [read file 1] [read file 2] > [outfile] • -R: indexes "read groups", required for GATK in later steps • e. g. : bwa mem -R '@RGt. ID: allt. SM: allt. LB: library' E_coli. fasta SRR 1770413_1. fastq SRR 1770413_2. fastq > align. sam

samtools: convert. sam to. bam, clean up names • . sam is the plain text output format of most sequence alignment programs • Because these can be large, most subsequent programs use the compressed ". bam" format instead • bwa sometimes does odd things to read pairing information, can clean up during conversion
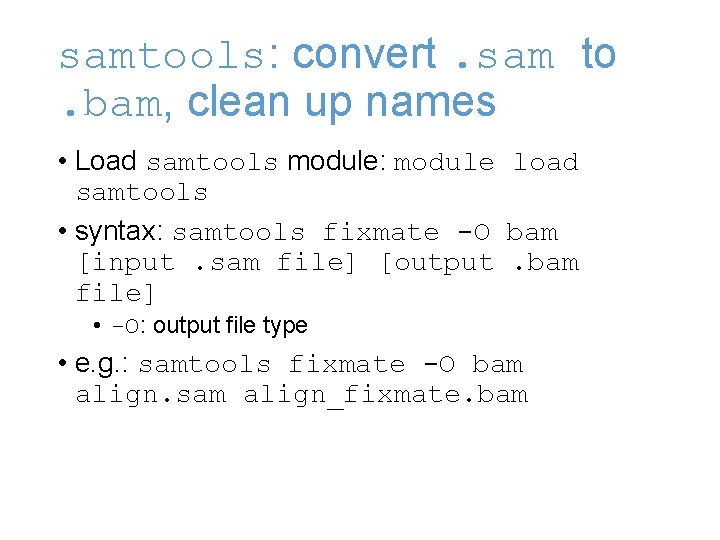
samtools: convert. sam to. bam, clean up names • Load samtools module: module load samtools • syntax: samtools fixmate -O bam [input. sam file] [output. bam file] • -O: output file type • e. g. : samtools fixmate -O bam align. sam align_fixmate. bam
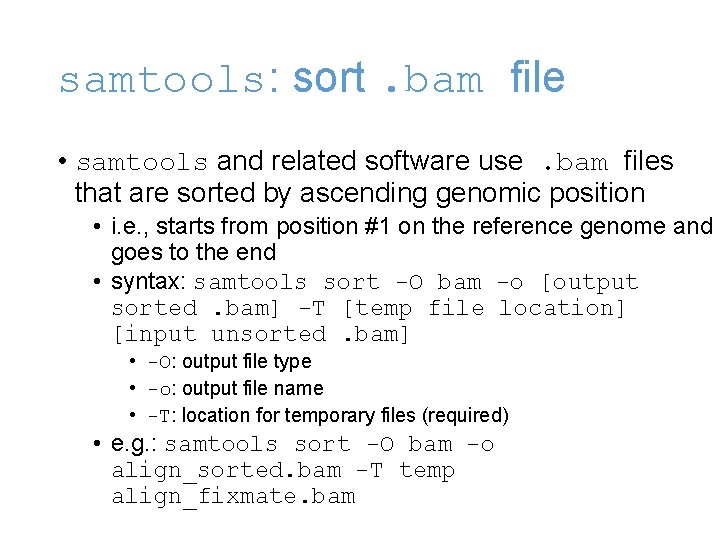
samtools: sort. bam file • samtools and related software use. bam files that are sorted by ascending genomic position • i. e. , starts from position #1 on the reference genome and goes to the end • syntax: samtools sort -O bam -o [output sorted. bam] -T [temp file location] [input unsorted. bam] • -O: output file type • -o: output file name • -T: location for temporary files (required) • e. g. : samtools sort -O bam -o align_sorted. bam -T temp align_fixmate. bam

GATK: realign indels • bwa sometimes misaligns indels in reads • One way to get rid of these is to use the realignment functions in the GATK package • More generally: GATK does much of the same thing as samtools, strong focus on diploid genomes • Unfortunately, GATK uses java (command line syntax) • Unfortunately, GATK needs its own file formats
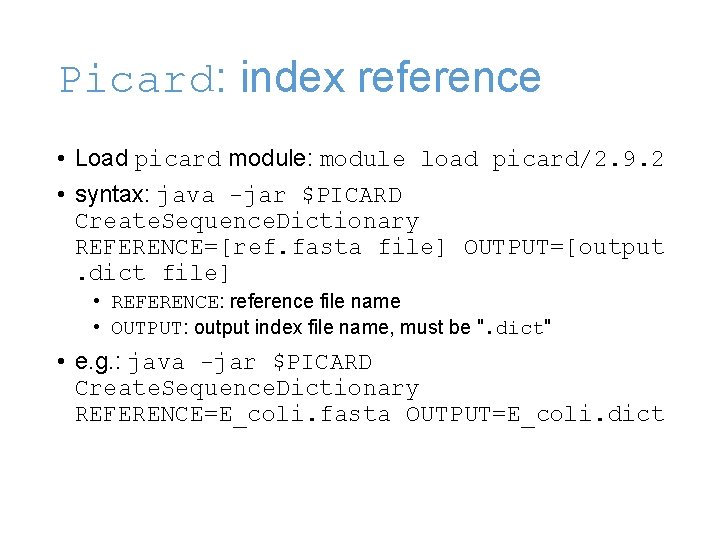
Picard: index reference • Load picard module: module load picard/2. 9. 2 • syntax: java -jar $PICARD Create. Sequence. Dictionary REFERENCE=[ref. fasta file] OUTPUT=[output. dict file] • REFERENCE: reference file name • OUTPUT: output index file name, must be ". dict" • e. g. : java -jar $PICARD Create. Sequence. Dictionary REFERENCE=E_coli. fasta OUTPUT=E_coli. dict
![samtools: index reference • syntax: samtools faidx [ref. fasta file] • e. g. : samtools: index reference • syntax: samtools faidx [ref. fasta file] • e. g. :](http://slidetodoc.com/presentation_image_h2/5eb34667466284a1c560d20f2d94989c/image-12.jpg)
samtools: index reference • syntax: samtools faidx [ref. fasta file] • e. g. : $ samtools faidx E_coli. fasta • outputs [ref. fasta]. fai index file
![samtools: index. bam file • syntax: samtools index [sorted. bam file] • e. g. samtools: index. bam file • syntax: samtools index [sorted. bam file] • e. g.](http://slidetodoc.com/presentation_image_h2/5eb34667466284a1c560d20f2d94989c/image-13.jpg)
samtools: index. bam file • syntax: samtools index [sorted. bam file] • e. g. : $ samtools index align_sorted. bam • outputs [sorted bam]. bai output file

GATK: prepare reads for indel realignment • Load GATK module: module load GATK/3. 7 • syntax: java -Xmx 2 g -jar $GATK -T Realigner. Target. Creator -R [ref. fasta file] -I [sorted & indexed. bam file] -o [output file name] • -R: reference. fasta file name • -I: sorted and indexed. bam file • -o: output intervals file name • e. g. : java -Xmx 2 g -jar $GATK -T Realigner. Target. Creator -R E_coli. fasta -I align_sorted. bam -o align_sorted. intervals
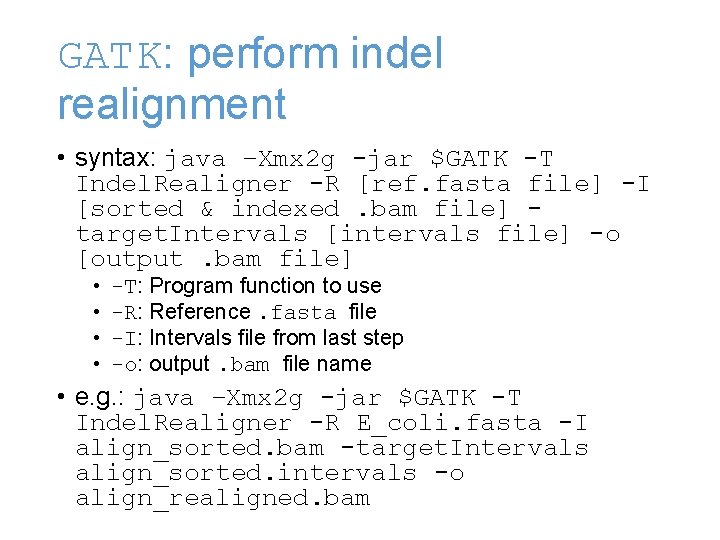
GATK: perform indel realignment • syntax: java –Xmx 2 g -jar $GATK -T Indel. Realigner -R [ref. fasta file] -I [sorted & indexed. bam file] target. Intervals [intervals file] -o [output. bam file] • • -T: Program function to use -R: Reference. fasta file -I: Intervals file from last step -o: output. bam file name • e. g. : java –Xmx 2 g -jar $GATK -T Indel. Realigner -R E_coli. fasta -I align_sorted. bam -target. Intervals align_sorted. intervals -o align_realigned. bam

Picard: remove duplicates • Duplicate reads can arise because of PCR artifacts during sequencing • Because duplicate reads to not provide additional information, it is best to remove them for computational efficiency • Identified by having identical start and end mapping positions

Picard: remove duplicates • syntax: java -Xmx 2 g -jar $PICARD Mark. Duplicates INPUT=[input bam file] OUTPUT=[output bam file] REMOVE_DUPLICATES=true METRICS_FILE=[metrics output file] • INPUT: input. bam file from GATK • OUTPUT: output. bam file lacking duplicates • METRICS_FILE: summary file of duplicate reads removed • e. g. : java -Xmx 2 g -jar $PICARD Mark. Duplicates INPUT=align_realigned. bam OUTPUT=align_nodups. bam REMOVE_DUPLICATES=true METRICS_FILE=nodups. metrics

samtools: index. bam file • samtools requires that the new. bam file be indexed before variant calling • syntax: samtools index [. bam file name] • e. g. : $ samtools index align_nodups. bam

samtools: create. bcf file for variant calling • bcftools is a package very similar to samtools that handles variant calling • Of course, it requires its own file format • syntax: samtools mpileup -go [output. bcf] -f [ref. fasta] [1 or more indexed. bam] • -go: specify output file name and. bcf format • -f: reference. fasta file name • e. g. : samtools mpileup -go E_coli. bcf -f E_coli. fasta align_nodups. bam
bcftools: call variants • The actual variant calling step uses the call function in bcftools • Load bcftools module: module load bcftools • syntax: bcftools call -vm. O z -o [output. vcf. gz file] [input. bcf file] • • -v: only output variant sites -m: specify variant calling algorithm (multiallelic) -O: specify output format, z =. vcf. gz -o: output file name • e. g. : bcftools call -vm. O z -o E_coli. vcf. gz E_coli. bcf

bcftools: index. vcf. gz file • tabix is a program included in bcftools that indexes a. vcf. gz file • Load tabix module: module load tabix • syntax: tabix -p vcf [input. vcf. gz file] • -p: specifies file type • e. g. : tabix -p vcf E_coli. vcf. gz
bcftools: analyze. vcf. gz file • bcftools handy software to analyze the variants that it has identified • syntax: bcftools stats -F [ref. fasta] -s - [input. vcf. gz file] > [output file] • -F: faidx indexed reference. fasta sequence • -s: list of samples to analyze, "-" = all samples • e. g. : bcftools stats -F E_coli. fasta -s - E_coli. vcf. gz > E_coli. vcf. gz. stats
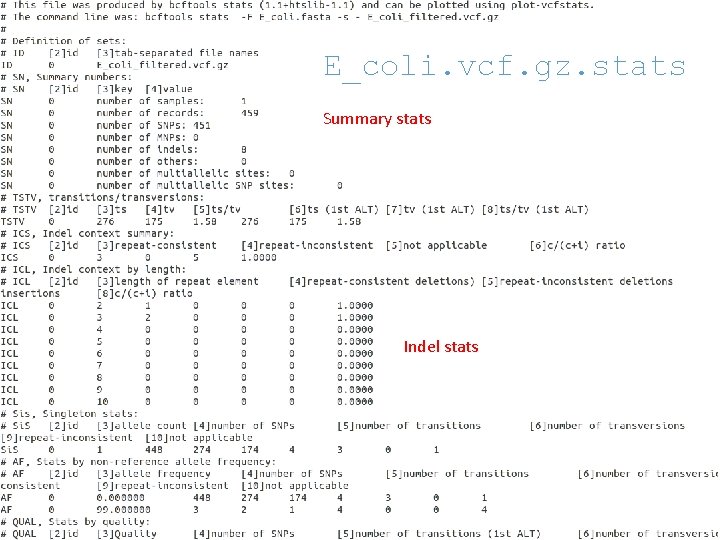
E_coli. vcf. gz. stats Summary stats Indel stats
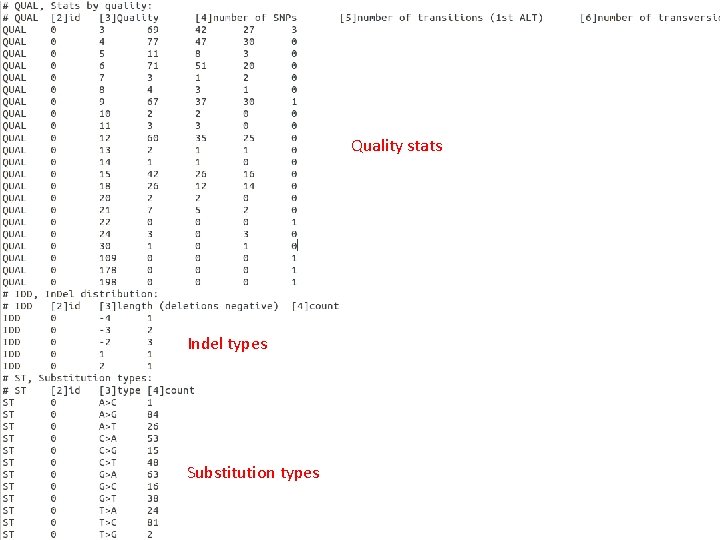
Quality stats Indel types Substitution types
bcftools: filter variants based on quality score • Generally, one wants to mark low quality variants. • How to draw a cutoff line is somewhat subjective • syntax: bcftools filter -O z -o [output. vcf. gz file] -s LOWQUAL -i '%QUAL>10' [input. vcf. gz file] • • -O: output type, "z" =. vcf. gz -o: output file name -s: label to mark failed variants -i: condition under which sequences pass • e. g. : bcftools filter -O z -o E_coli_filtered. vcf. gz -s LOWQUAL -i '%QUAL>10' E_coli. vcf. gz
bcftools: calculate stats based on filtered variants • You can tell bcftools stats to only analyze variants that pass the filter • syntax: bcftools stats -F [ref. fasta] -f PASS -s - [input filtered. vcf. gz file] • -F: faidx indexed reference. fasta sequence • -f: how sequences to include are marked • -s: list of samples to analyze, "-" = all samples • e. g. : bcftools stats -F E_coli. fasta -f PASS -s - E_coli_filtered. vcf. gz > E_coli. vcf. gz. stats

Exercise #13 • Download the complete E. coli B str. REL 606 genome as your reference strain • Determine how the E. coli K-12 strain sequenced in SRR 1770413 differs from REL 606 1. Use head to make test datasets 2. Run each command individually, pasting successful commands into a sbatch script 3. Run entire sbatch script on test data 4. Run entire sbatch script on real data • NOTE: each run of the pipeline needs to be in its own directory with only the input files in it
- Slides: 27