Maximize read usage through mapping strategies Key concepts











- Slides: 27

Maximize read usage through mapping strategies

Key concepts of session • The settings you use MATTER. • More reads mapping is not ALWAYS better. • Sometimes you need to understand what you threw away. • Mapping is yet another quality control step.

What are we doing with mapping? How do we align the bag of reads to the reference? – Efficiently (memory and time) – Account for inexact matches and ambiguity? Traditional sequence alignment (BLAT, Blast) are too slow for millions of short reads.

Short read mapping • Input: – A reference genome – A collection of many 25 -250 bp tags/reads – User-specified parameters • Output: – One or more genomic coordinates for each tag • In practice, not all reads successfully map to the reference genome. Why?

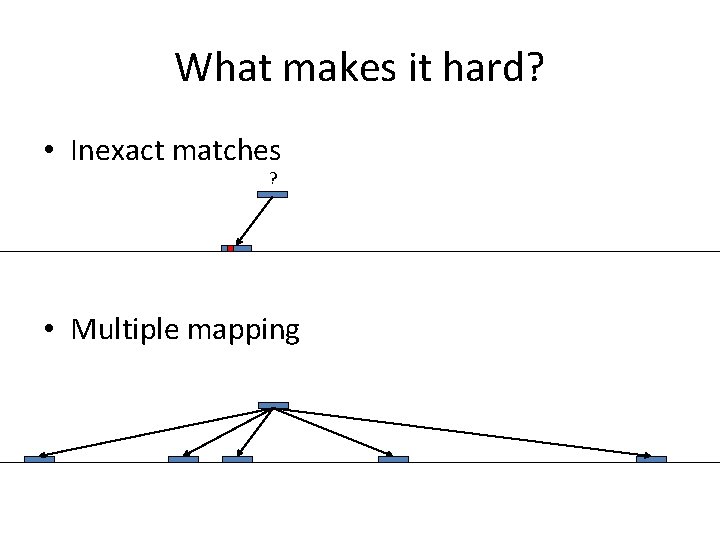
What makes it hard? • Inexact matches ? • Multiple mapping

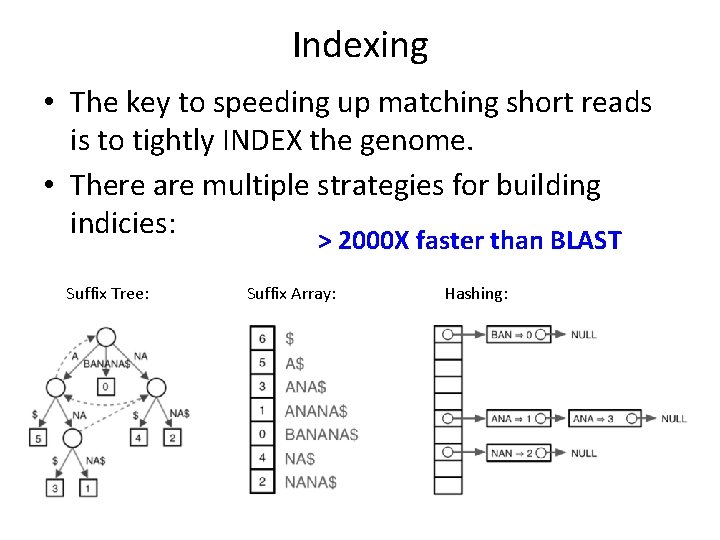
Indexing • The key to speeding up matching short reads is to tightly INDEX the genome. • There are multiple strategies for building indicies: > 2000 X faster than BLAST Suffix Tree: Suffix Array: Hashing:

Bowtie • Indexes the reference genome using a scheme based on the Burrows-Wheeler transform (BWT) which is very space efficient. • A quality-aware backtracking algorithm that allows mismatches and favors high-quality alignments. • Double indexing', a strategy to avoid excessive backtracking

Bowtie caveat “If one or more exact matches exist for a read, then Bowtie is guaranteed to report one, but if the best match is an inexact one then Bowtie is not guaranteed in all cases to find the highest quality alignment. ” …unless you use the MUCH slower “best” option

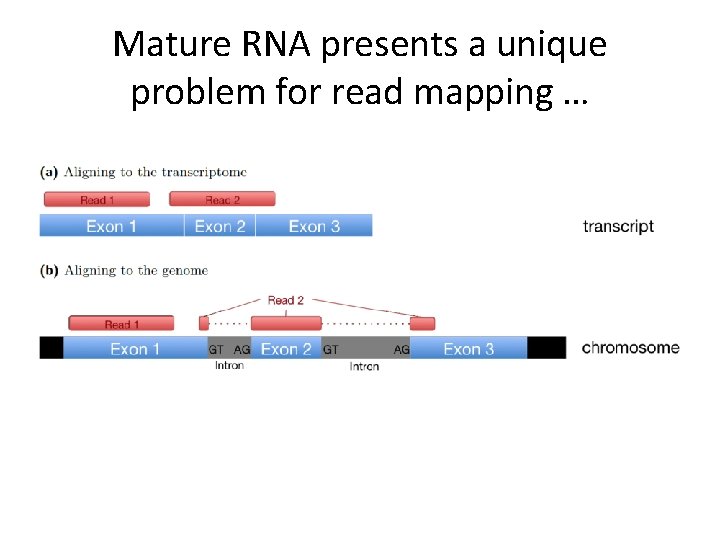
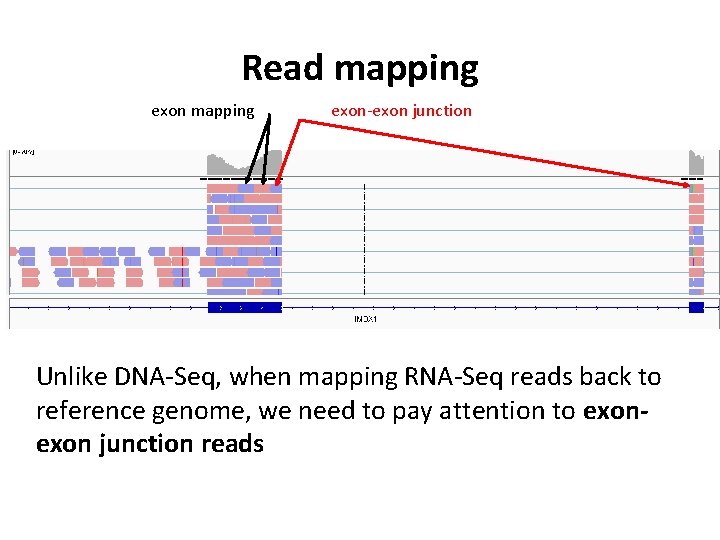
Mature RNA presents a unique problem for read mapping …

Read mapping exon-exon junction Unlike DNA-Seq, when mapping RNA-Seq reads back to reference genome, we need to pay attention to exon junction reads

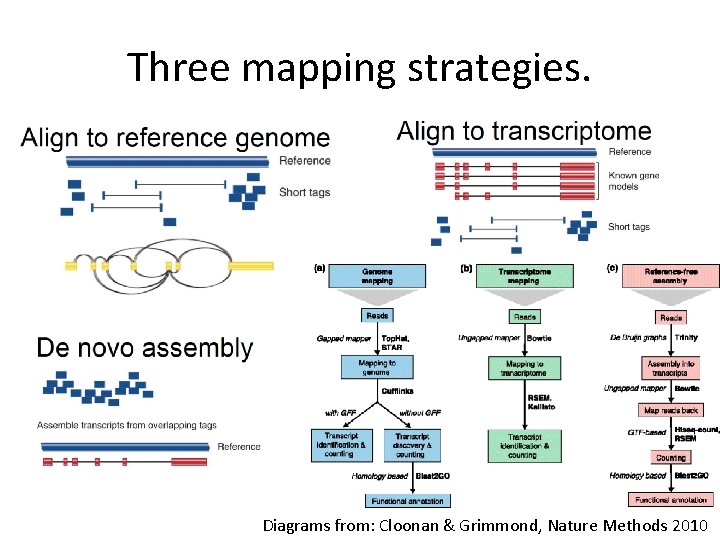
Three mapping strategies. Diagrams from: Cloonan & Grimmond, Nature Methods 2010

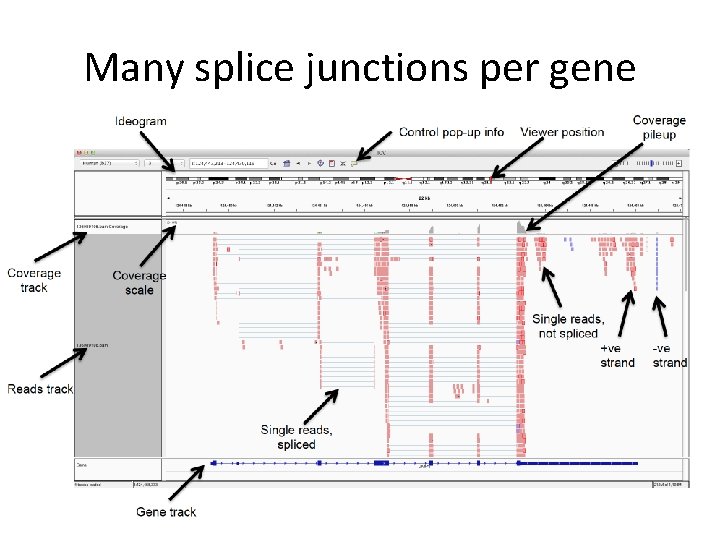
Many splice junctions per gene

Mapping software …

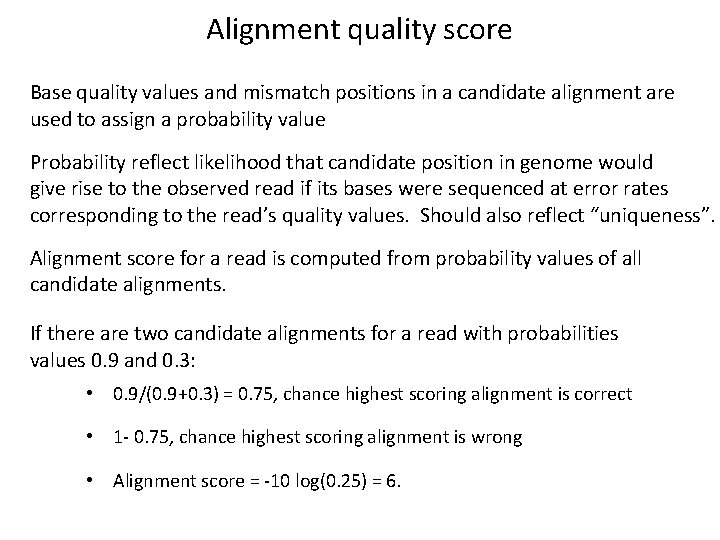
Alignment quality score Base quality values and mismatch positions in a candidate alignment are used to assign a probability value Probability reflect likelihood that candidate position in genome would give rise to the observed read if its bases were sequenced at error rates corresponding to the read’s quality values. Should also reflect “uniqueness”. Alignment score for a read is computed from probability values of all candidate alignments. If there are two candidate alignments for a read with probabilities values 0. 9 and 0. 3: • 0. 9/(0. 9+0. 3) = 0. 75, chance highest scoring alignment is correct • 1 - 0. 75, chance highest scoring alignment is wrong • Alignment score = -10 log(0. 25) = 6.

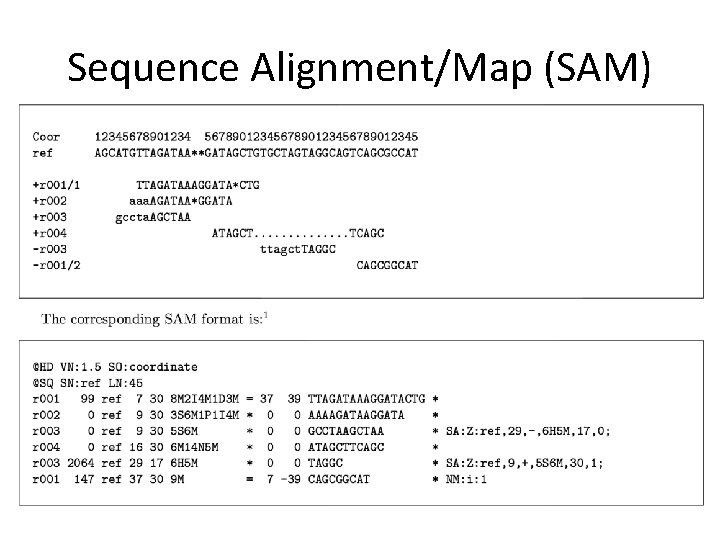
Sequence Alignment/Map (SAM)

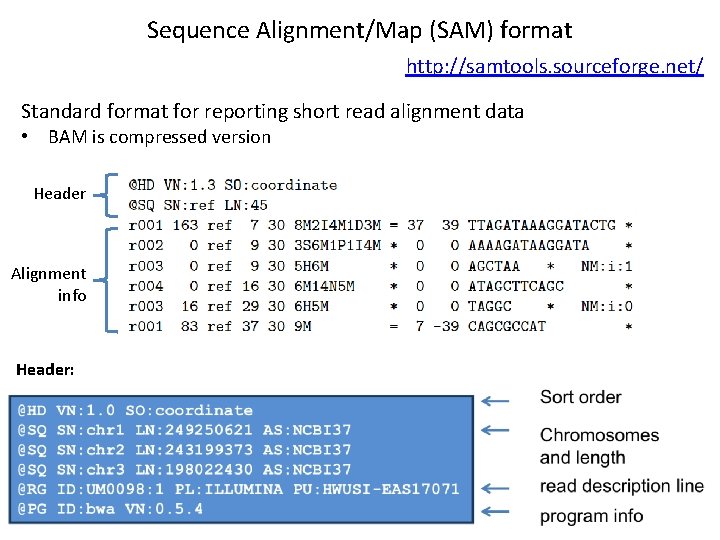
Sequence Alignment/Map (SAM) format http: //samtools. sourceforge. net/ Standard format for reporting short read alignment data • BAM is compressed version Header Alignment info Header:

Read Information

Read Information

Mapping Flags

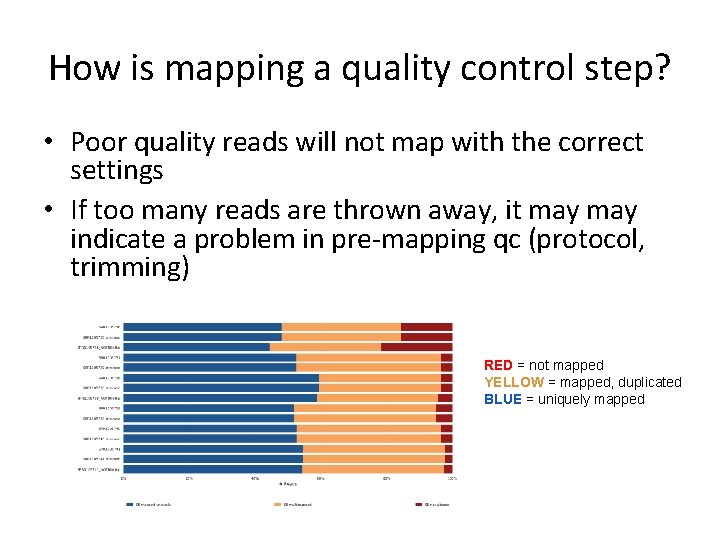
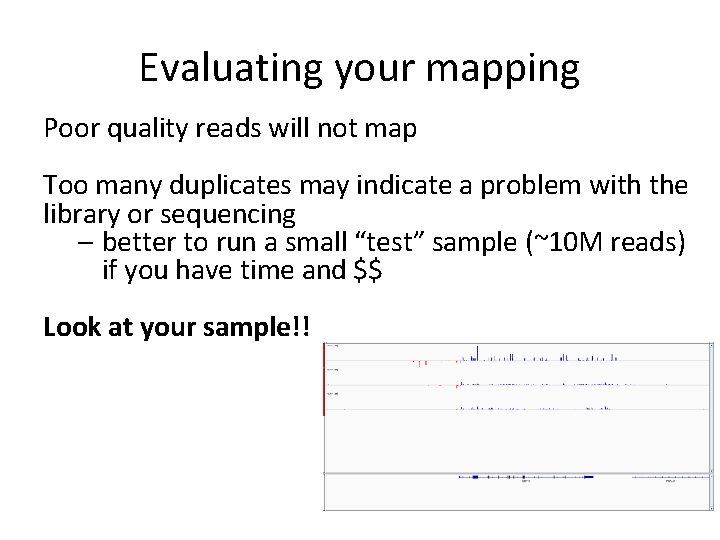
How is mapping a quality control step? • Poor quality reads will not map with the correct settings • If too many reads are thrown away, it may indicate a problem in pre-mapping qc (protocol, trimming) RED = not mapped YELLOW = mapped, duplicated BLUE = uniquely mapped

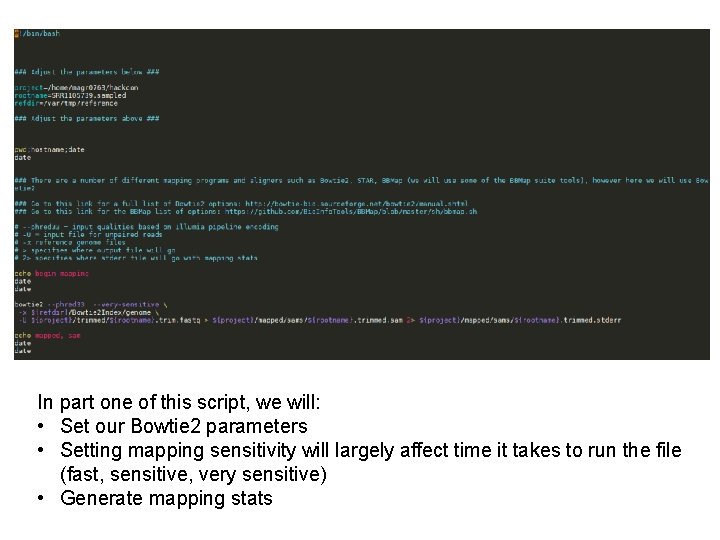
Mapping : from fastq to tdf • After successful trimming, it’s time to map • Again, there a number of different mapping tools all with pros and cons • We will use Bowtie 2

In part one of this script, we will: • Set our Bowtie 2 parameters • Setting mapping sensitivity will largely affect time it takes to run the file (fast, sensitive, very sensitive) • Generate mapping stats

In the next part of this script, we will: • Convert. sam to. bam (binary sam, quicker processing) and index see handout • Generate bed. Graph file which gives information about read coverage

Lastly, we will: • Read count correct – adjust for read depth post-mapping using flagstat file • Convert. bed. Graph to. tdf for rapid loading into IGV To run (after adjusting rootname/project – refdir is the same for everyone): $ bash mapping. sh

Open tdfs in IGV Path = $ cd data/nascent-ws/Mapped/tdfs/ Open all SRRs with. tri. tdf : see dowell. colorad. edu/Hack. Con/pages/visualiza tion. html for IGV tips and tricks

Evaluating your mapping Poor quality reads will not map Too many duplicates may indicate a problem with the library or sequencing – better to run a small “test” sample (~10 M reads) if you have time and $$ Look at your sample!!

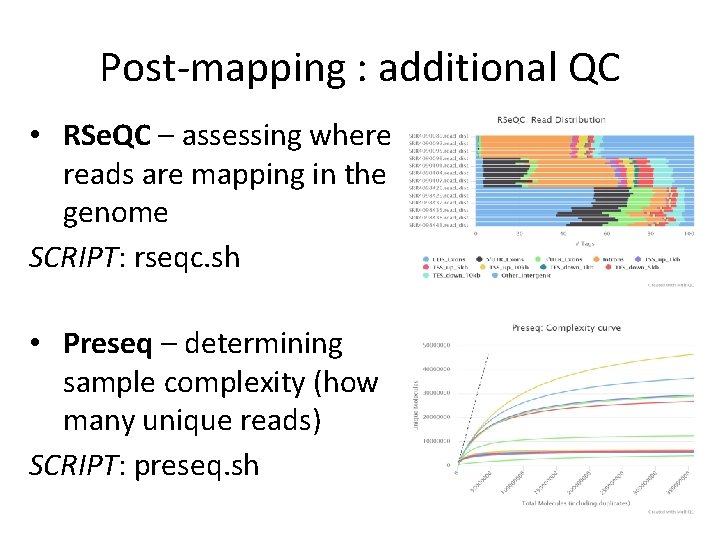
Post-mapping : additional QC • RSe. QC – assessing where reads are mapping in the genome SCRIPT: rseqc. sh • Preseq – determining sample complexity (how many unique reads) SCRIPT: preseq. sh