Max Planck Institute for Molecular Genetics A pipeline




- Slides: 19

Max Planck Institute for Molecular Genetics A pipeline based on multivariate correspondence analysis with supplementary variables for cancer genomics Christine Steinhoff Max Planck Institute for Molecular Genetics Berlin, Germany

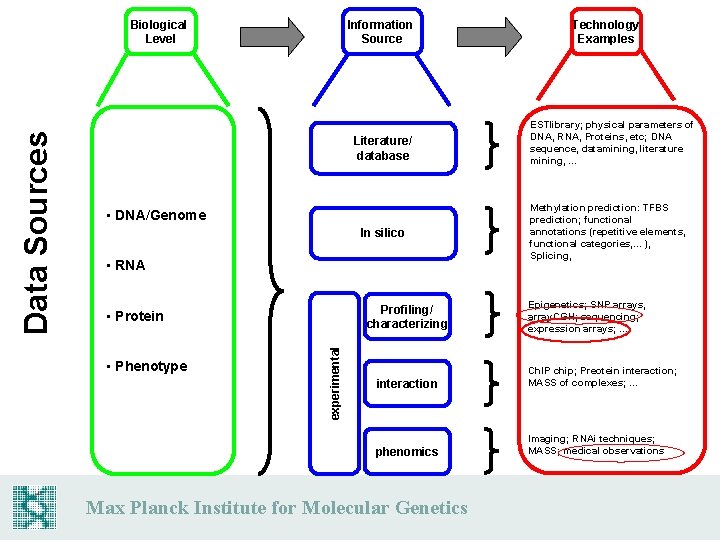
Information Source Literature/ database • DNA/Genome In silico • RNA Profiling/ characterizing • Protein • Phenotype experimental Data Sources Biological Level Technology Examples ESTlibrary; physical parameters of DNA, RNA, Proteins, etc; DNA sequence, datamining, literature mining, . . . Methylation prediction: TFBS prediction; functional annotations (repetitive elements, functional categories, . . . ), Splicing, Epigenetics; SNP arrays, array. CGH; sequencing; expression arrays; . . . interaction Ch. IP chip; Preotein interaction; MASS of complexes; . . . phenomics Imaging; RNAi techniques; MASS; medical observations Max Planck Institute for Molecular Genetics

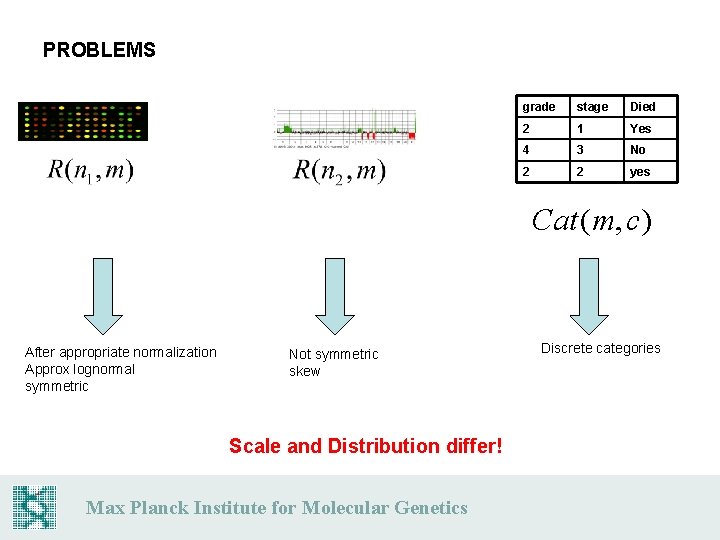
PROBLEMS grade stage Died 2 1 Yes 4 3 No 2 2 yes Cat (m, c) After appropriate normalization Approx lognormal symmetric Not symmetric skew Scale and Distribution differ! Max Planck Institute for Molecular Genetics Discrete categories

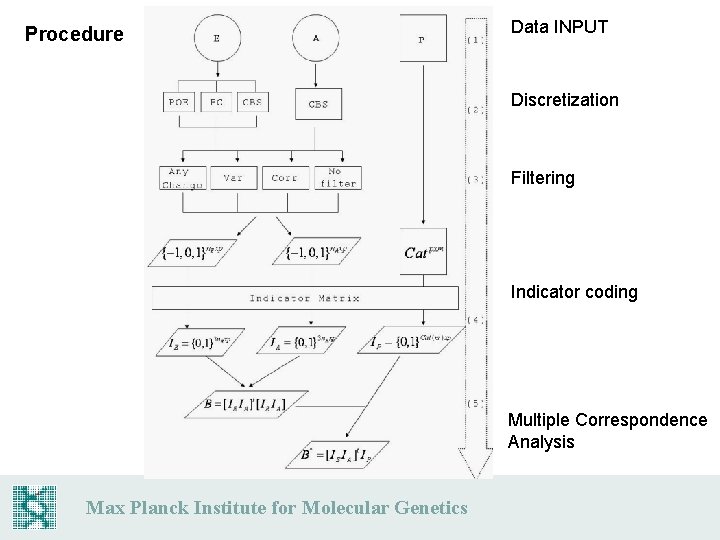
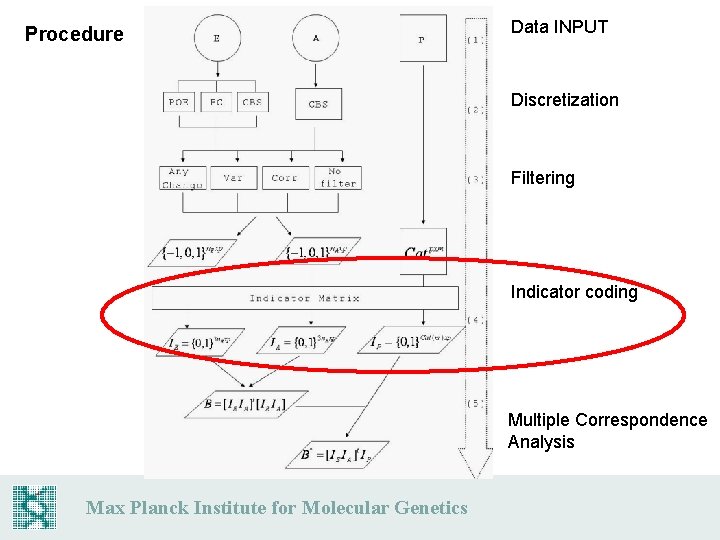
Procedure Data INPUT Discretization Filtering Indicator coding Multiple Correspondence Analysis Max Planck Institute for Molecular Genetics

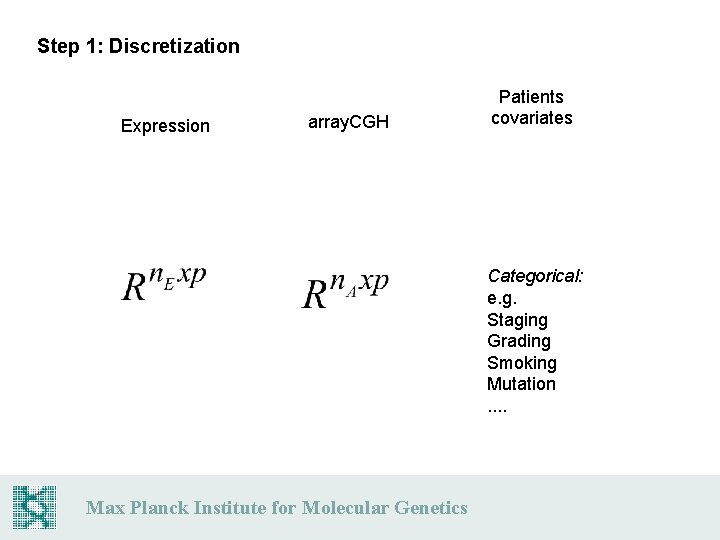
Step 1: Discretization Expression array. CGH Patients covariates Categorical: e. g. Staging Grading Smoking Mutation. . Max Planck Institute for Molecular Genetics

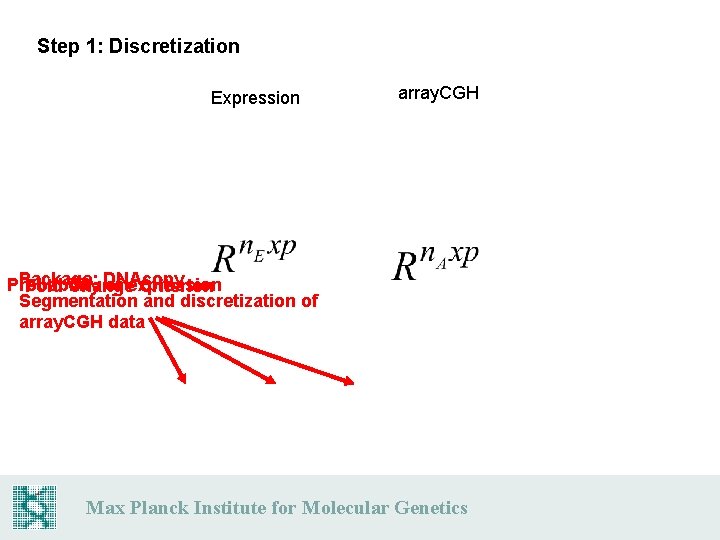
Step 1: Discretization Expression array. CGH Package: DNAcopy Probability of expression Fold Change Criterion Segmentation and discretization of array. CGH data Max Planck Institute for Molecular Genetics

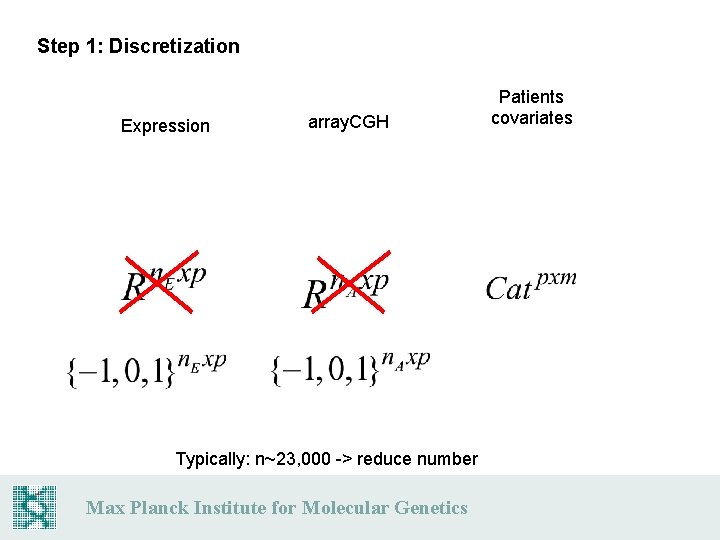
Step 1: Discretization Expression array. CGH Typically: n~23, 000 -> reduce number Max Planck Institute for Molecular Genetics Patients covariates

Step 2: Filtering (optional) Possibilities -Neglect all genes with no change in any patient -Choose genes with highest Variance across patients -Select for high Correlation between array. CGH and expression Max Planck Institute for Molecular Genetics

Procedure Data INPUT Discretization Filtering Indicator coding Multiple Correspondence Analysis Max Planck Institute for Molecular Genetics

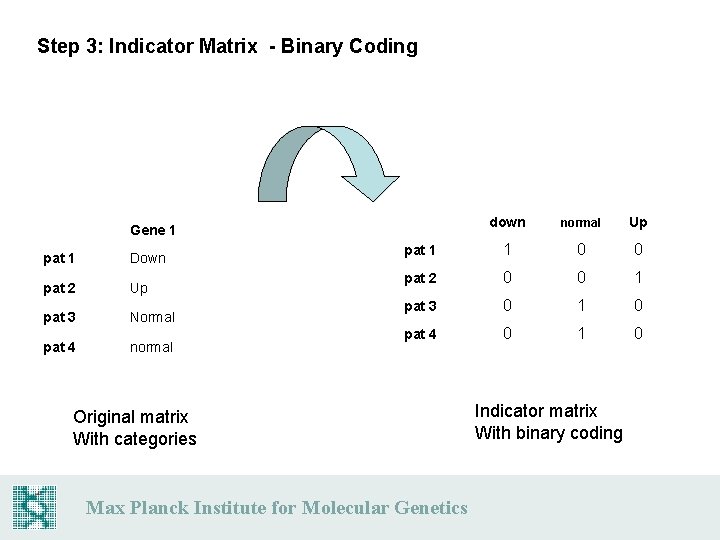
Step 3: Indicator Matrix - Binary Coding down normal Up pat 1 1 0 0 pat 2 0 0 1 pat 3 0 1 0 pat 4 0 1 0 Gene 1 pat 1 Down pat 2 Up pat 3 Normal pat 4 normal Original matrix With categories Max Planck Institute for Molecular Genetics Indicator matrix With binary coding

From: Multiple Correspondence Analysis and related Methods Max Planck Institute for Molecular Genetics

EXAMPLE: PUBLISHED DATA Max Planck Institute for Molecular Genetics

Covariate States‘ Display Max Planck Institute for Molecular Genetics

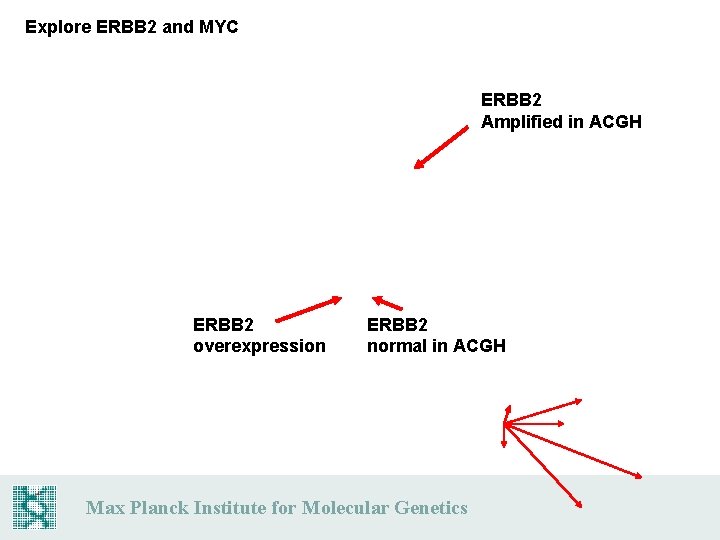
Explore ERBB 2 and MYC ERBB 2 Amplified in ACGH ERBB 2 overexpression ERBB 2 normal in ACGH Max Planck Institute for Molecular Genetics

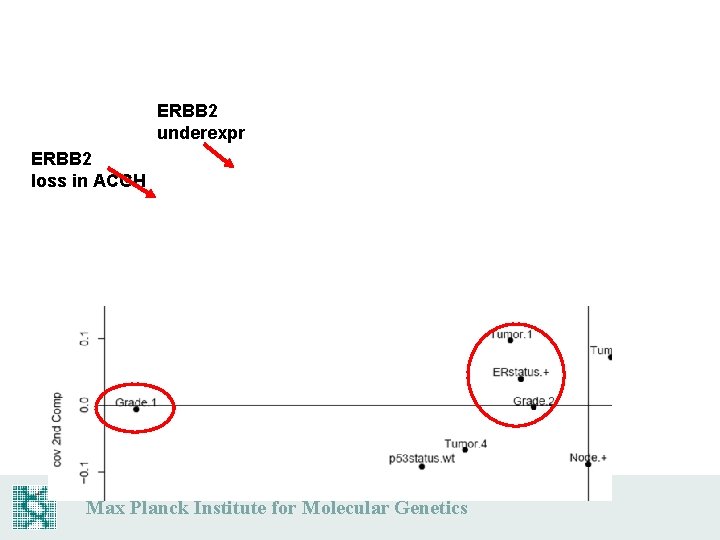
ERBB 2 underexpr ERBB 2 loss in ACGH Max Planck Institute for Molecular Genetics

MYC amplification MYC Overexpression Max Planck Institute for Molecular Genetics

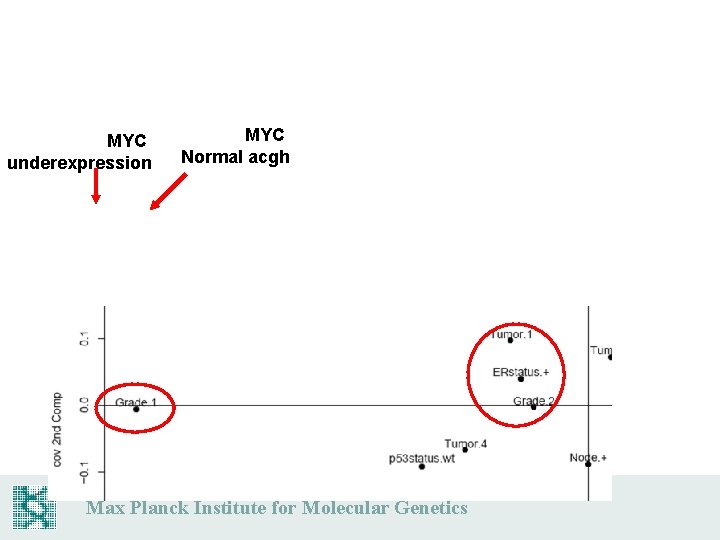
MYC underexpression MYC Normal acgh Max Planck Institute for Molecular Genetics

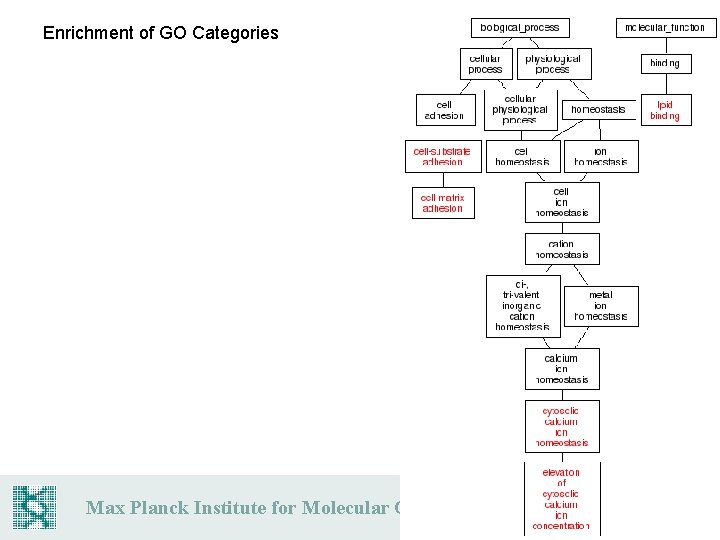
Enrichment of GO Categories Max Planck Institute for Molecular Genetics

Thank you for your attention ! ACKNOWLEDGEMENT Max Planck Institute for Molecular Genetics Martin Vingron Sensor Lab, CNR-INFM Matteo Pardo Max Planck Institute for Molecular Genetics
 Max planck institute neuroscience internship
Max planck institute neuroscience internship Max planck
Max planck 1900 max planck
1900 max planck Max planck
Max planck Gyrokinetic
Gyrokinetic Modelo atomico de max planck
Modelo atomico de max planck Max planck institut rechtsgeschichte
Max planck institut rechtsgeschichte Max planck encyclopedia of comparative constitutional law
Max planck encyclopedia of comparative constitutional law Bert l. de groot
Bert l. de groot Max planck
Max planck Difference between linear pipeline and non linear pipeline
Difference between linear pipeline and non linear pipeline Superscalar pipeline design
Superscalar pipeline design 14-3 human molecular genetics
14-3 human molecular genetics Golg11
Golg11 Chapter 12 section 1 molecular genetics answer key
Chapter 12 section 1 molecular genetics answer key Molecular genetics section 1 dna the genetic material
Molecular genetics section 1 dna the genetic material Mega molecular
Mega molecular Chapter 12 section 1 dna the genetic material
Chapter 12 section 1 dna the genetic material Molecular genetics chapter 12
Molecular genetics chapter 12 Parent genotype
Parent genotype