Mass Spectrometrybased Proteomics SANOVO CONFIDENTIAL boydfuturist New methodology
Mass Spectrometry-based Proteomics

SANOVO CONFIDENTIAL boydfuturist
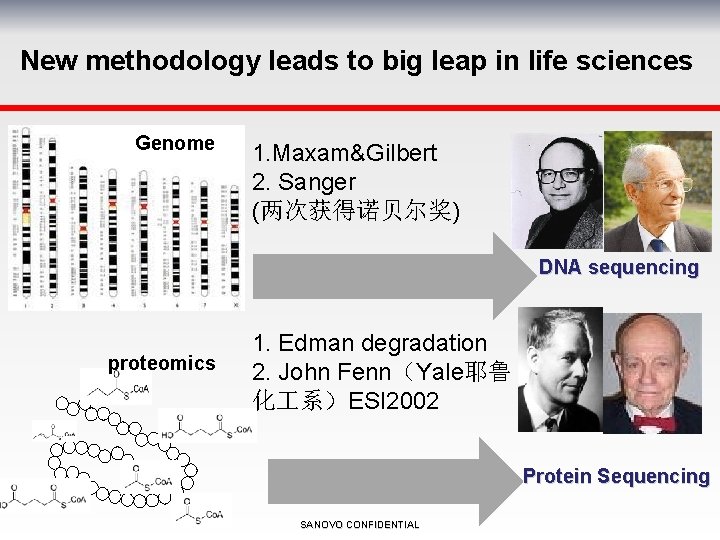
New methodology leads to big leap in life sciences Genome 1. Maxam&Gilbert 2. Sanger (两次获得诺贝尔奖) DNA sequencing proteomics 1. Edman degradation 2. John Fenn(Yale耶鲁 化 系)ESI 2002 Protein Sequencing SANOVO CONFIDENTIAL

LC-MS and LC-MS/MS Protein Trypsin-digested peptides Protein pattern info Peptide sequence info Should contain the information for the genome code and the epi-proteomics code ? ? ? Liquid-Chromatography (LC) MS (mass of peptides) MS/MS (breakdown of peptide)
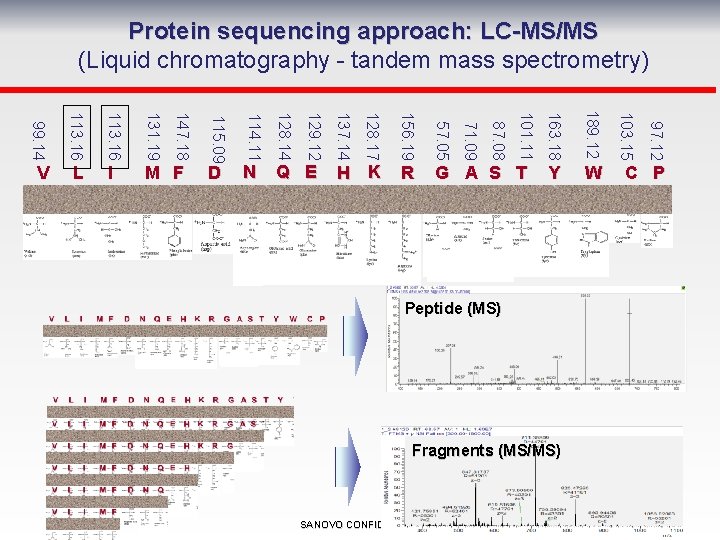
Protein sequencing approach: LC-MS/MS (Liquid chromatography - tandem mass spectrometry) W Peptide (MS) Fragments (MS/MS) SANOVO CONFIDENTIAL 97. 12 189. 12 Y 103. 15 163. 18 G A S T 101. 11 R 87. 08 57. 05 H K 71. 09 156. 19 Q E 128. 17 N 137. 14 D 129. 12 M F 128. 14 131. 19 I 114. 11 113. 16 L 115. 09 113. 16 147. 18 99. 14 V C P
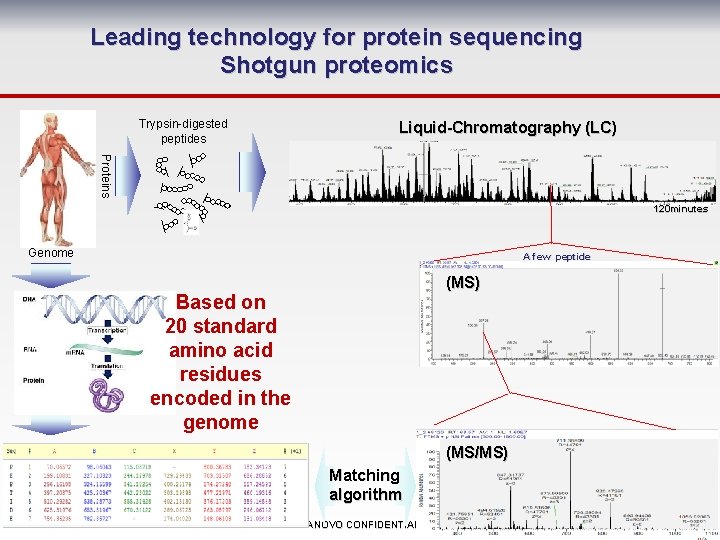
Leading technology for protein sequencing Shotgun proteomics Trypsin-digested peptides Liquid-Chromatography (LC) Proteins 120 minutes Genome A few peptide (MS) Based on 20 standard amino acid residues encoded in the genome (MS/MS) Matching algorithm SANOVO CONFIDENTIAL

How to sequence a peptide? P | EPTIDE PE | PTIDE theoretical PEP | TIDE PEPT | IDE PEPTID | E Orbitrap MS/MS SANOVO CONFIDENTIAL
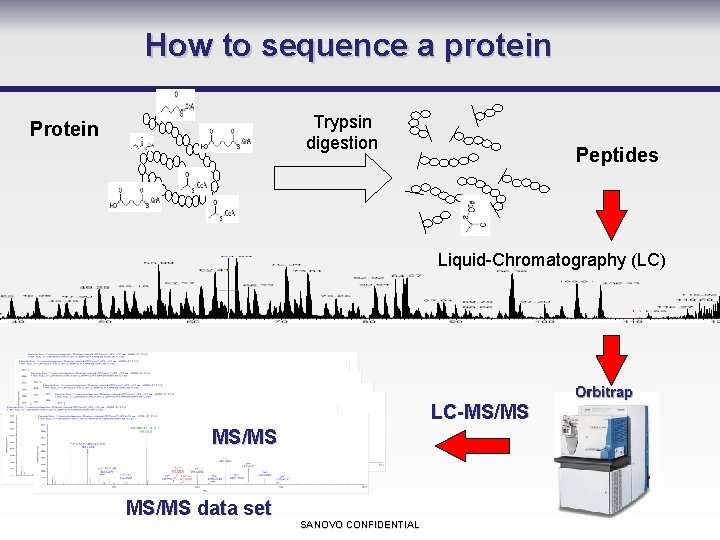
How to sequence a protein Trypsin digestion Protein Peptides Liquid-Chromatography (LC) LC-MS/MS data set SANOVO CONFIDENTIAL

Example : sequencing of a single protein sequencing of GFAP from post mortem brain 1 51 101 151 201 251 301 351 401 MERRRITSAA DFSLAGALNA LNQLRAKEPT VRQKLQDETN RKIHEEEVRE HEAEEWYRSK GTNESLERQM DLLNVKLALD EGHLKRNIVV RRSYVSSGEM GFKETRASER KLADVYQAEL LRLEAENNLA LQEQLARQQV FADLTDAAAR REQEERHVRE IEIATYRKLL KTVEMRDGEV MVGGLAPGRR AEMMELNDRF RELRLRLDQL AYRQEADEAT HVELDVAKPD NAELLRQAKH AASYQEALAR EGEENRITIP IKESKQEHKD LGPGTRLSLA ASYIEKVRFL TANSARLEVE LARLDLERKI LTAALKEIRT EANDYRRQLQ LEEEGQSLKD VQTFSNLQIR VM SANOVO CONFIDENTIAL RMPPPLPTRV EQQNKALAAE RDNLAQDLAT ESLEEEIRFL QYEAMASSNM SLTCDLESLR EMARHLQEYQ ETSLDTKSVS

Proteomics of human diseases SANOVO CONFIDENTIAL
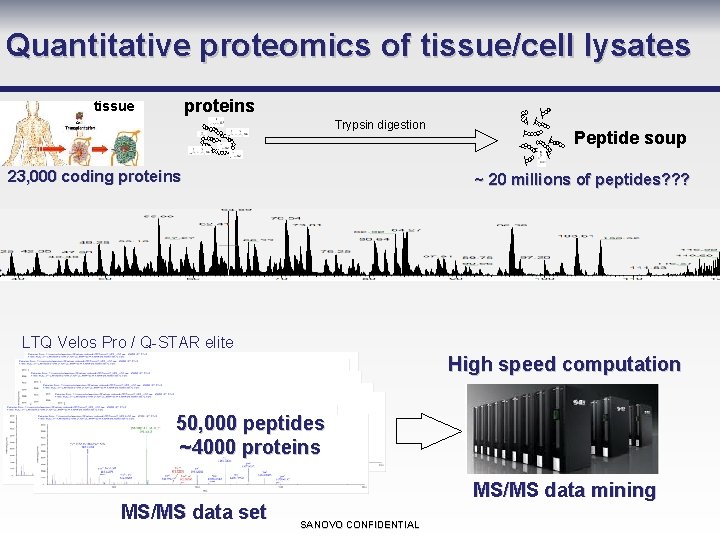
Quantitative proteomics of tissue/cell lysates proteins tissue Trypsin digestion 23, 000 coding proteins Peptide soup ~ 20 millions of peptides? ? ? LTQ Velos Pro / Q-STAR elite High speed computation 50, 000 peptides ~4000 proteins MS/MS data set MS/MS data mining SANOVO CONFIDENTIAL
Proteomics of human cancers Human Pancreatic cancer/adjacent Human glioma tumor stage I/IV Human kidney tumor/adjacent Human gastric cancer//adjacent Mn. SOD Oxidative Modification Mouse Oxidative Stress SANOVO CONFIDENTIAL
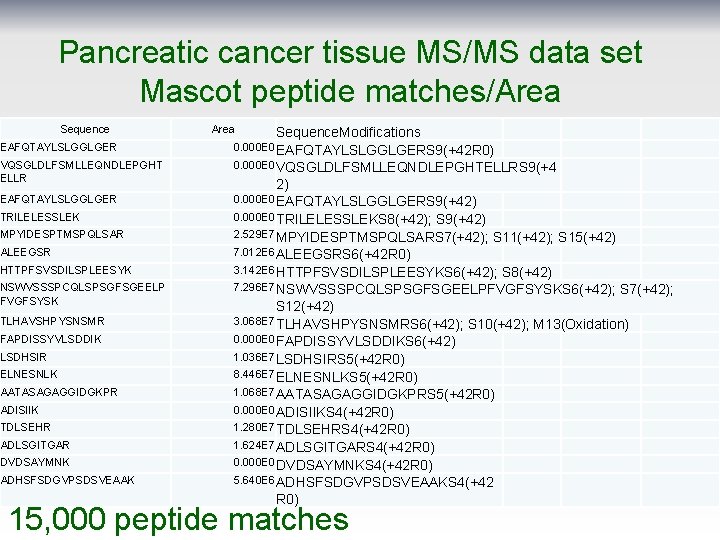
Pancreatic cancer tissue MS/MS data set Mascot peptide matches/Area Sequence. Modifications EAFQTAYLSLGGLGER 0. 000 E 0 EAFQTAYLSLGGLGERS 9(+42 R 0) VQSGLDLFSMLLEQNDLEPGHT ELLR 0. 000 E 0 VQSGLDLFSMLLEQNDLEPGHTELLRS 9(+4 EAFQTAYLSLGGLGER 0. 000 E 0 EAFQTAYLSLGGLGERS 9(+42) TRILELESSLEK 0. 000 E 0 TRILELESSLEKS 8(+42); MPYIDESPTMSPQLSAR 2. 529 E 7 MPYIDESPTMSPQLSARS 7(+42); ALEEGSR 7. 012 E 6 ALEEGSRS 6(+42 R 0) HTTPFSVSDILSPLEESYK 3. 142 E 6 HTTPFSVSDILSPLEESYKS 6(+42); NSWVSSSPCQLSPSGFSGEELP FVGFSYSK 7. 296 E 7 NSWVSSSPCQLSPSGFSGEELPFVGFSYSKS 6(+42); TLHAVSHPYSNSMR FAPDISSYVLSDDIK LSDHSIR ELNESNLK AATASAGAGGIDGKPR ADISIIK TDLSEHR ADLSGITGAR DVDSAYMNK ADHSFSDGVPSDSVEAAK 2) S 9(+42) S 11(+42); S 15(+42) S 8(+42) S 7(+42); S 12(+42) 3. 068 E 7 TLHAVSHPYSNSMRS 6(+42); S 10(+42); M 13(Oxidation) 0. 000 E 0 FAPDISSYVLSDDIKS 6(+42) 1. 036 E 7 LSDHSIRS 5(+42 R 0) 8. 446 E 7 ELNESNLKS 5(+42 R 0) 1. 068 E 7 AATASAGAGGIDGKPRS 5(+42 R 0) 0. 000 E 0 ADISIIKS 4(+42 R 0) 1. 280 E 7 TDLSEHRS 4(+42 R 0) 1. 624 E 7 ADLSGITGARS 4(+42 R 0) 0. 000 E 0 DVDSAYMNKS 4(+42 R 0) 5. 640 E 6 ADHSFSDGVPSDSVEAAKS 4(+42 R 0) 15, 000 peptide matches
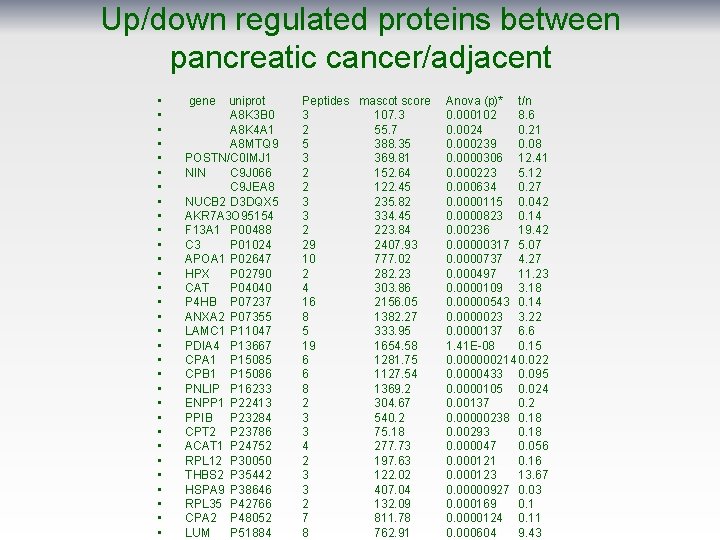
Up/down regulated proteins between pancreatic cancer/adjacent • • • • • • • • gene uniprot A 8 K 3 B 0 A 8 K 4 A 1 A 8 MTQ 9 POSTN/C 0 IMJ 1 NIN C 9 J 066 C 9 JEA 8 NUCB 2 D 3 DQX 5 AKR 7 A 3 O 95154 F 13 A 1 P 00488 C 3 P 01024 APOA 1 P 02647 HPX P 02790 CAT P 04040 P 4 HB P 07237 ANXA 2 P 07355 LAMC 1 P 11047 PDIA 4 P 13667 CPA 1 P 15085 CPB 1 P 15086 PNLIP P 16233 ENPP 1 P 22413 PPIB P 23284 CPT 2 P 23786 ACAT 1 P 24752 RPL 12 P 30050 THBS 2 P 35442 HSPA 9 P 38646 RPL 35 P 42766 CPA 2 P 48052 LUM P 51884 Peptides mascot score 3 107. 3 2 55. 7 5 388. 35 3 369. 81 2 152. 64 2 122. 45 3 235. 82 3 334. 45 2 223. 84 29 2407. 93 10 777. 02 2 282. 23 4 303. 86 16 2156. 05 8 1382. 27 5 333. 95 19 1654. 58 6 1281. 75 6 1127. 54 8 1369. 2 2 304. 67 3 540. 2 3 75. 18 4 277. 73 2 197. 63 3 122. 02 3 407. 04 2 132. 09 7 811. 78 8 762. 91 Anova (p)* t/n 0. 000102 8. 6 0. 0024 0. 21 0. 000239 0. 08 0. 0000306 12. 41 0. 000223 5. 12 0. 000634 0. 27 0. 0000115 0. 042 0. 0000823 0. 14 0. 00236 19. 42 0. 00000317 5. 07 0. 0000737 4. 27 0. 000497 11. 23 0. 0000109 3. 18 0. 00000543 0. 14 0. 0000023 3. 22 0. 0000137 6. 6 1. 41 E-08 0. 15 0. 000000214 0. 022 0. 0000433 0. 095 0. 0000105 0. 024 0. 00137 0. 2 0. 00000238 0. 18 0. 00293 0. 18 0. 000047 0. 056 0. 000121 0. 16 0. 000123 13. 67 0. 00000927 0. 03 0. 000169 0. 1 0. 0000124 0. 11 0. 000604 9. 43
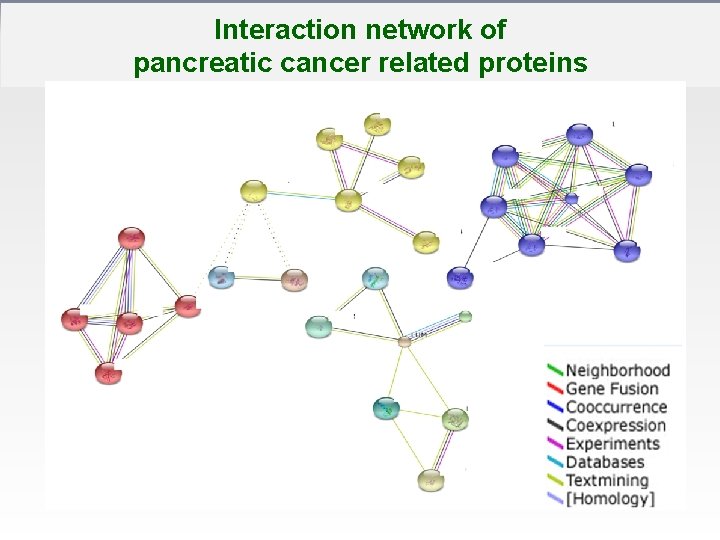
Interaction network of pancreatic cancer related proteins
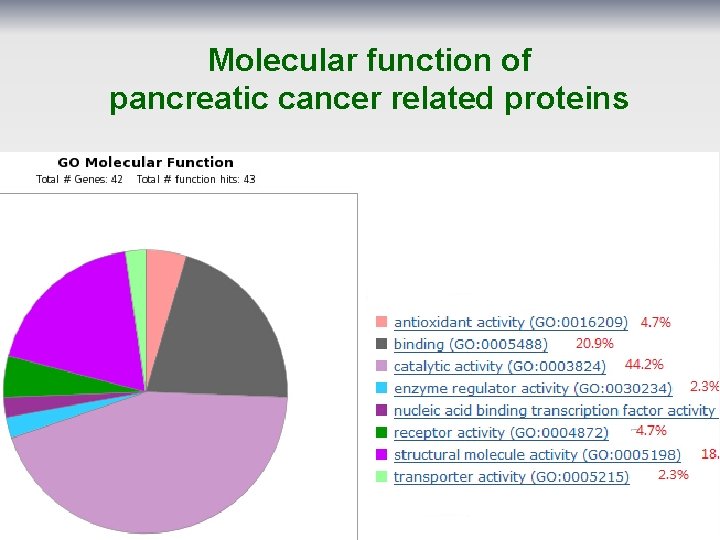
Molecular function of pancreatic cancer related proteins
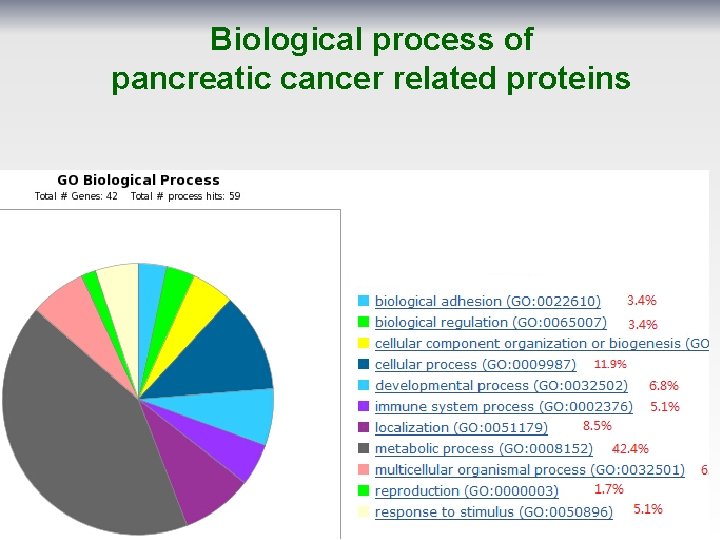
Biological process of pancreatic cancer related proteins
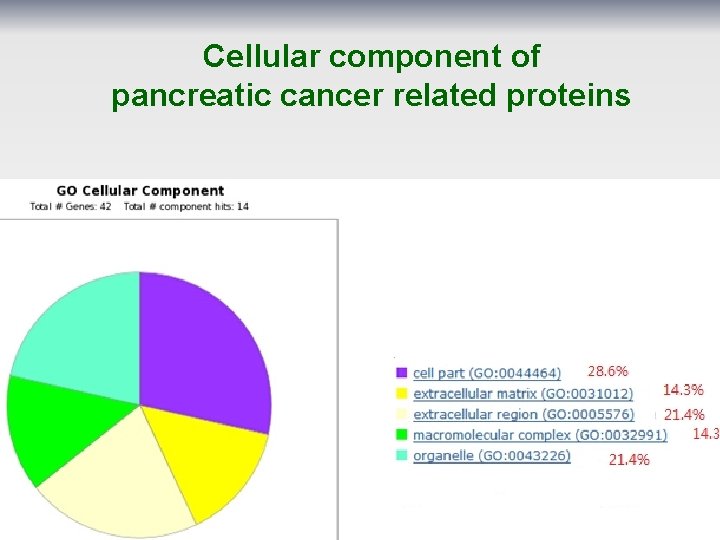
Cellular component of pancreatic cancer related proteins
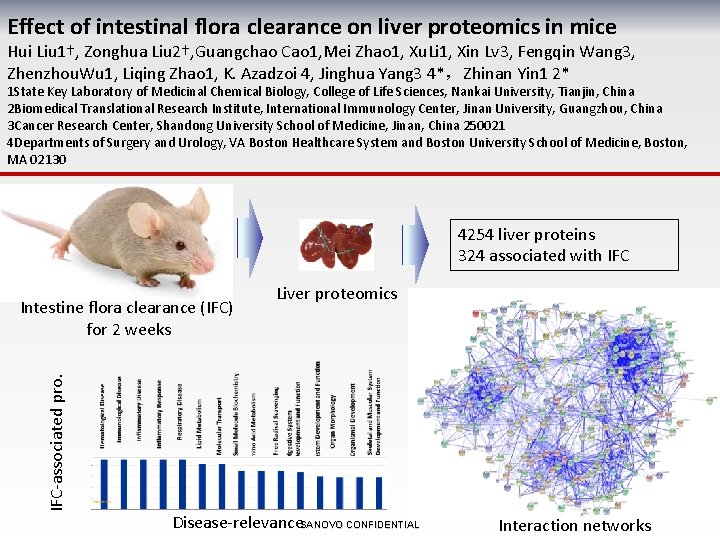
Effect of intestinal flora clearance on liver proteomics in mice Hui Liu 1†, Zonghua Liu 2†, Guangchao Cao 1, Mei Zhao 1, Xu. Li 1, Xin Lv 3, Fengqin Wang 3, Zhenzhou. Wu 1, Liqing Zhao 1, K. Azadzoi 4, Jinghua Yang 3 4*,Zhinan Yin 1 2* 1 State Key Laboratory of Medicinal Chemical Biology, College of Life Sciences, Nankai University, Tianjin, China 2 Biomedical Translational Research Institute, International Immunology Center, Jinan University, Guangzhou, China 3 Cancer Research Center, Shandong University School of Medicine, Jinan, China 250021 4 Departments of Surgery and Urology, VA Boston Healthcare System and Boston University School of Medicine, Boston, MA 02130 4254 liver proteins 324 associated with IFC-associated pro. Intestine flora clearance (IFC) for 2 weeks Liver proteomics Disease-relevance. SANOVO CONFIDENTIAL Interaction networks
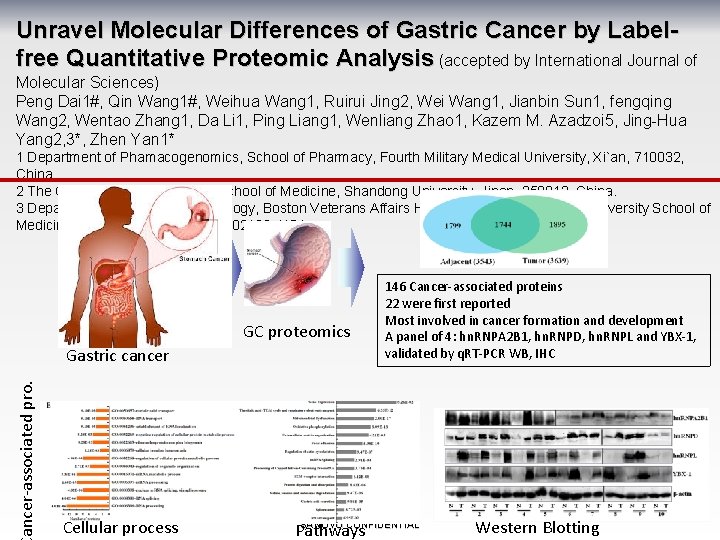
Unravel Molecular Differences of Gastric Cancer by Labelfree Quantitative Proteomic Analysis (accepted by International Journal of Molecular Sciences) Peng Dai 1#, Qin Wang 1#, Weihua Wang 1, Ruirui Jing 2, Wei Wang 1, Jianbin Sun 1, fengqing Wang 2, Wentao Zhang 1, Da Li 1, Ping Liang 1, Wenliang Zhao 1, Kazem M. Azadzoi 5, Jing-Hua Yang 2, 3*, Zhen Yan 1* 1 Department of Phamacogenomics, School of Pharmacy, Fourth Military Medical University, Xi`an, 710032, China. 2 The Cancer Research Center, School of Medicine, Shandong University, Jinan, 250012, China. 3 Departments of Surgery and Urology, Boston Veterans Affairs Healthcare System, Boston University School of Medicine, Boston, Massachusetts, 02130, USA. GC proteomics ancer-associated pro. Gastric cancer Cellular process 146 Cancer-associated proteins 22 were first reported Most involved in cancer formation and development A panel of 4: hn. RNPA 2 B 1, hn. RNPD, hn. RNPL and YBX-1, validated by q. RT-PCR WB, IHC SANOVO CONFIDENTIAL Pathways Western Blotting
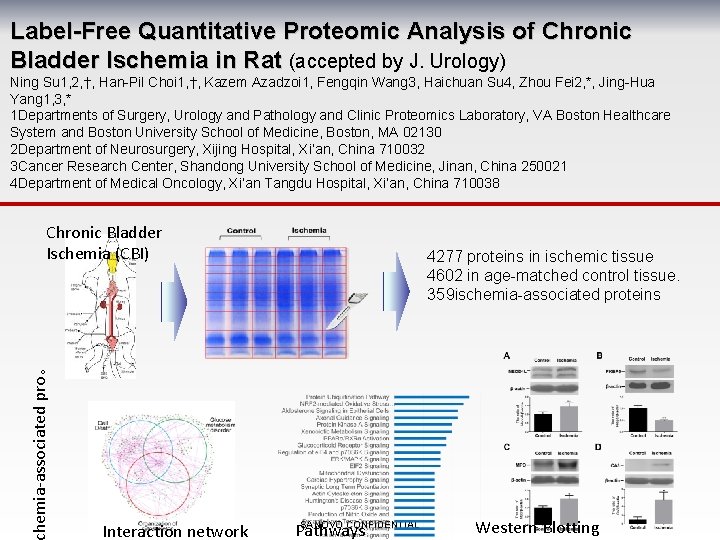
Label-Free Quantitative Proteomic Analysis of Chronic Bladder Ischemia in Rat (accepted by J. Urology) Ning Su 1, 2, †, Han-Pil Choi 1, †, Kazem Azadzoi 1, Fengqin Wang 3, Haichuan Su 4, Zhou Fei 2, *, Jing-Hua Yang 1, 3, * 1 Departments of Surgery, Urology and Pathology and Clinic Proteomics Laboratory, VA Boston Healthcare System and Boston University School of Medicine, Boston, MA 02130 2 Department of Neurosurgery, Xijing Hospital, Xi’an, China 710032 3 Cancer Research Center, Shandong University School of Medicine, Jinan, China 250021 4 Department of Medical Oncology, Xi’an Tangdu Hospital, Xi’an, China 710038 hemia-associated pro。 Chronic Bladder Ischemia (CBI) Interaction network 4277 proteins in ischemic tissue 4602 in age-matched control tissue. 359 ischemia-associated proteins SANOVO CONFIDENTIAL Pathways Western Blotting

Homework www. course. sdu. edu. cn/G 2 S/pm. cc 1. Manually compare the intensities (area) of stomach cancer and adjacent tissues by label-free quantification (Sequest Search datasets are provided through We. Chat group) 2. Dai, P. , et al. "Unraveling Molecular Differences of Gastric Cancer by Label-Free Quantitative Proteomics Analysis. " International Journal of Molecular Sciences 17. 2(2015). 3. Su N, Choi H P, Wang F, et al. Quantitative proteomic analysis of differentially expressed proteins and downstream signaling pathways in chronic bladder ischemia. [J]. Journal of Urology, 2015, 195(2): 515 -523. < Epiproteomics> Class 2017
- Slides: 22