Mass spectrometry is an analytical technique that can





















- Slides: 34

Mass spectrometry is an analytical technique that can be used to deduce the molecular formula of an unknown compound. Gaseous molecules of the compound are bombarded with high-speed electrons from an electron gun. These knock out an electron from some of the molecules, creating molecular ions (M+), which travel to the M(g) + e- M+(g) + 2 edetector plates: The relative abundances of the detected ions form a mass spectrum: a kind of molecular fingerprint that can be identified by computer using a spectral database. 1 of 45 © Boardworks Ltd 2010

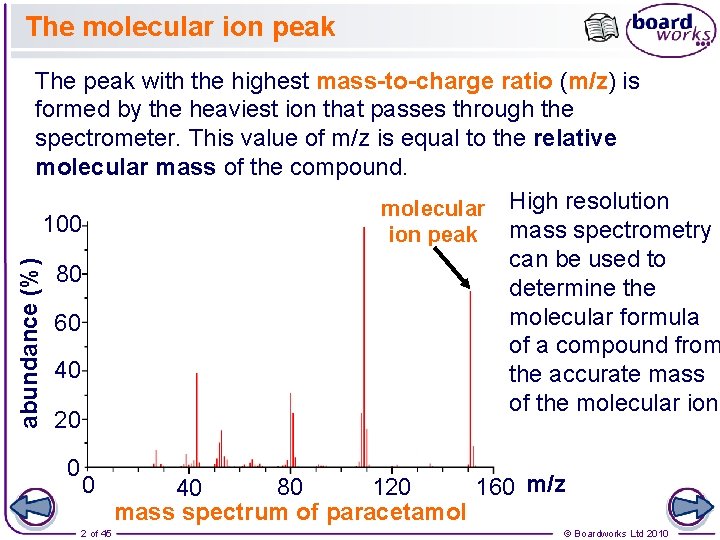
The molecular ion peak abundance (%) The peak with the highest mass-to-charge ratio (m/z) is formed by the heaviest ion that passes through the spectrometer. This value of m/z is equal to the relative molecular mass of the compound. molecular High resolution 100 ion peak mass spectrometry can be used to 80 determine the molecular formula 60 of a compound from 40 the accurate mass of the molecular ion. 20 0 0 2 of 45 160 m/z 80 120 40 mass spectrum of paracetamol © Boardworks Ltd 2010

What is fragmentation? A molecular ion is a positively-charged ion, which is also a radical as it contains a single unpaired electron. It is therefore sometimes represented as M+ • . During mass spectroscopy, the molecular ion can fragment into a positive ion and a radical: M+ • → X+ + Y • For example, in the case of propane: or CH 3 CH 2 CH 3+ • → CH 3 CH 2+ + CH 3 • CH 3 CH 2 CH 3+ • → CH 3 CH 2 • + CH 3+ NB: Only the ions are detected by the mass spectrometer. This fragmentation process gives rise to characteristic peaks on a mass spectrum that can give information about the structure of the molecule. 3 of 45 © Boardworks Ltd 2010

Fragmentation and molecular structure 4 of 45 © Boardworks Ltd 2010

Fragmentation of carbonyl compounds 5 of 45 © Boardworks Ltd 2010

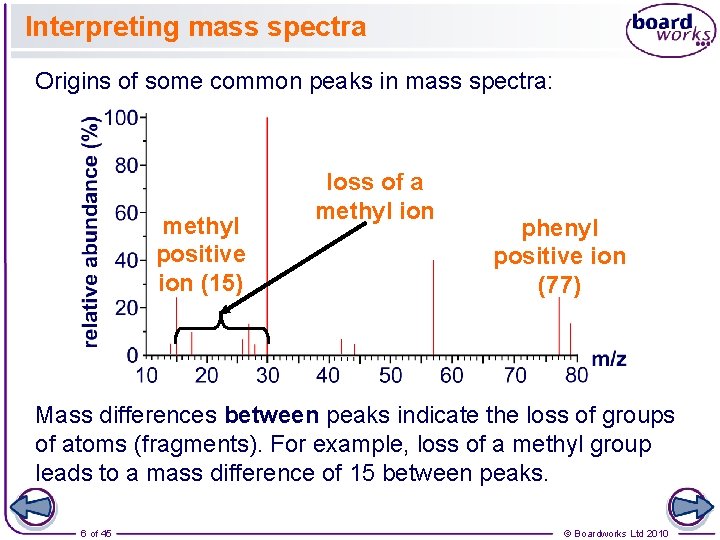
Interpreting mass spectra Origins of some common peaks in mass spectra: methyl positive ion (15) loss of a methyl ion phenyl positive ion (77) Mass differences between peaks indicate the loss of groups of atoms (fragments). For example, loss of a methyl group leads to a mass difference of 15 between peaks. 6 of 45 © Boardworks Ltd 2010

Interpreting mass spectra activity 7 of 45 © Boardworks Ltd 2010

Uses of mass spectrometry Some uses of mass spectrometry: l Identifying elements in new or foreign substances, for example analysing samples from the Mars space probe. l Monitoring levels of environmental pollution, for example amounts of lead or pesticide in a sample. l In biochemical research, for example determining the composition of a protein by comparing it against a database of known compounds. 8 of 45 © Boardworks Ltd 2010

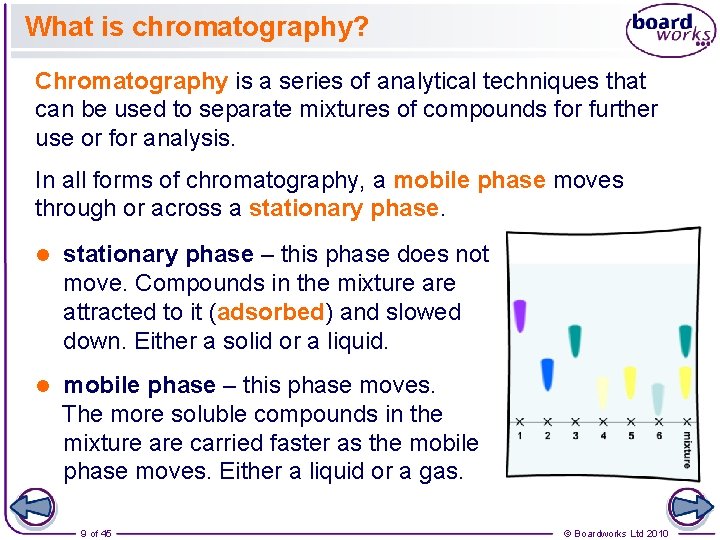
What is chromatography? Chromatography is a series of analytical techniques that can be used to separate mixtures of compounds for further use or for analysis. In all forms of chromatography, a mobile phase moves through or across a stationary phase. l stationary phase – this phase does not move. Compounds in the mixture attracted to it (adsorbed) and slowed down. Either a solid or a liquid. l mobile phase – this phase moves. The more soluble compounds in the mixture are carried faster as the mobile phase moves. Either a liquid or a gas. 9 of 45 © Boardworks Ltd 2010

Thin layer chromatography 10 of 45 © Boardworks Ltd 2010

Column chromatography 11 of 45 © Boardworks Ltd 2010

Gas–liquid chromatography 12 of 45 © Boardworks Ltd 2010

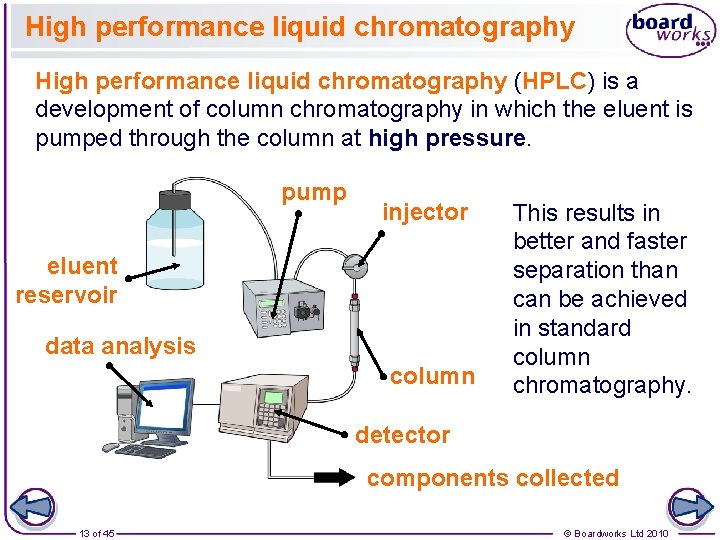
High performance liquid chromatography (HPLC) is a development of column chromatography in which the eluent is pumped through the column at high pressure. pump injector eluent reservoir data analysis column This results in better and faster separation than can be achieved in standard column chromatography. detector components collected 13 of 45 © Boardworks Ltd 2010

Gas chromatography–mass spectroscopy In both GL chromatography and HPLC, the output from the chromatography column can be passed through a mass spectrometer. The spectra obtained can be compared to spectra of known compounds. Gas chromatography– mass spectroscopy (GC –MS) is used extensively in forensics, environmental monitoring and in airport security systems. It is sensitive enough to detect minute quantities of substances 14 of 45 © Boardworks Ltd 2010

Chromatography: true or false? 15 of 45 © Boardworks Ltd 2010

Glossary 16 of 45 © Boardworks Ltd 2010

What’s the keyword? 17 of 45 © Boardworks Ltd 2010

Multiple-choice quiz 18 of 45 © Boardworks Ltd 2010

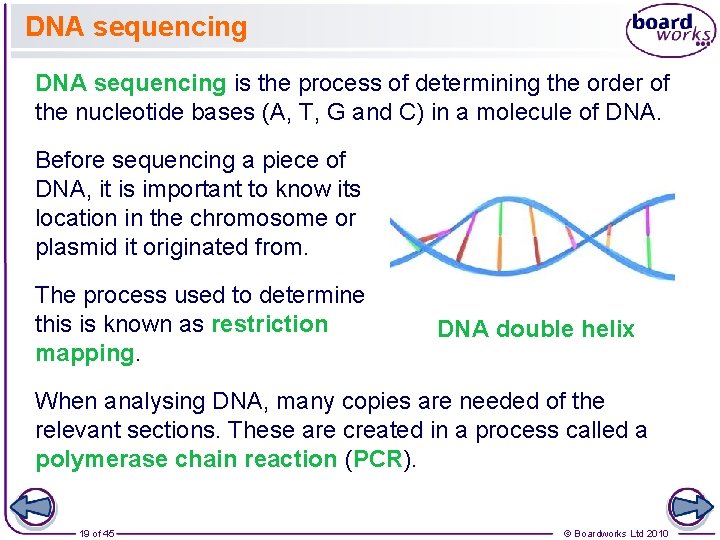
DNA sequencing is the process of determining the order of the nucleotide bases (A, T, G and C) in a molecule of DNA. Before sequencing a piece of DNA, it is important to know its location in the chromosome or plasmid it originated from. The process used to determine this is known as restriction mapping. DNA double helix When analysing DNA, many copies are needed of the relevant sections. These are created in a process called a polymerase chain reaction (PCR). 19 of 45 © Boardworks Ltd 2010

Producing multiple copies of DNA 20 of 45 © Boardworks Ltd 2010

Locating genes 21 of 45 © Boardworks Ltd 2010

The Sanger method 22 of 45 © Boardworks Ltd 2010

PCR – the basic ingredients 23 of 45 © Boardworks Ltd 2010

Studying genes – true or false 24 of 45 © Boardworks Ltd 2010

Materials used in DNA sequencing 25 of 45 © Boardworks Ltd 2010

26 of 45 © Boardworks Ltd 2010

Full genome sequencing involves sequencing not only nuclear DNA, but also the DNA contained within mitochondria and chloroplasts. With this vast quantity of information, comparisons can be made between individuals of the same species and between different species. This gives us insights into evolutionary relationships, and differing responses to medical treatments. 27 of 45 © Boardworks Ltd 2010

Minisatellites 28 of 45 © Boardworks Ltd 2010

Genetic fingerprinting A genetic fingerprint is created by looking at an individual’s minisatellites. The choice of minisatellite is very important: the profiler needs to choose those minisatellites that show the most variation, to reduce the chance that two individuals could have matching fingerprints. The main stages involved in genetic fingerprinting are: 1. Extraction 2. Digestion 3. Separation 4. Hybridization 5. Development 29 of 45 © Boardworks Ltd 2010

Genetic fingerprinting 30 of 45 © Boardworks Ltd 2010

Analysing genetic fingerprints A profiler can inspect genetic fingerprints by eye to make quick comparisons. This can be a useful tool in forensic science. The process can also be automated with a computer using the marker bands to calculate the size and distance travelled by the bands in the profile. It is sometimes necessary to consider the odds that somebody else in the population has the same DNA fingerprint as the one being studied. For instance, to assess the risk of a false criminal conviction. 31 of 45 © Boardworks Ltd 2010

The Human Genome Project (HGP) was an international, publicly funded venture to sequence three billion bases in the 20, 000– 25, 000 genes of the human genome. The project ran from 1990 to 2003, when a first full sequence was published. Corrections and further analyses have been added to the results over the following years, and are still being added today. One of the subsidiary aims of the HGP was to study the ethical and social implications of the project, and to predict and discuss the legal issues that would arise. Can you think of any examples? 32 of 45 © Boardworks Ltd 2010

Designer drugs One of the outcomes of the HGP is the development of new drugs. Drugs can be designed using the knowledge of protein structure, gained from the gene sequence information. This reduces the need for trial and error and allows doctors to tailor specific treatments to their patients, depending on the exact nature of their condition. Designer drugs can be used to treat the fundamental causes of a disease rather than just the symptoms. 33 of 45 © Boardworks Ltd 2010

How is a DNA profile created? 34 of 45 © Boardworks Ltd 2010