Mass Spectrometry 101 An Introductory Lecture On Mass










- Slides: 58

Mass Spectrometry 101 An Introductory Lecture On Mass Spectrometry Fundamentals Presented to the Sandler Mass Spectrometry Users’ Group University of California San Francisco April 11, 2003

What does a mass spectrometer do? 1. It measures mass better than any other technique. 2. It can give information about chemical structures. What are mass measurements good for? To identify, verify, and quantitate: metabolites, recombinant proteins, proteins isolated from natural sources, oligonucleotides, drug candidates, peptides, synthetic organic chemicals, polymers

Applications of Mass Spectrometry Pharmaceutical analysis Bioavailability studies Drug metabolism studies, pharmacokinetics Characterization of potential drugs Drug degradation product analysis Screening of drug candidates Identifying drug targets Biomolecule characterization Proteins and peptides Oligonucleotides Environmental analysis Pesticides on foods Soil and groundwater contamination Forensic analysis/clinical

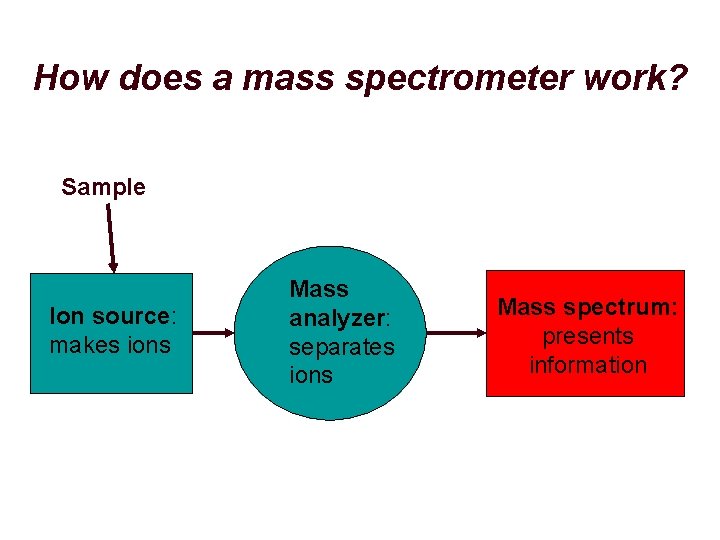
How does a mass spectrometer work? Sample Ion source: makes ions Mass analyzer: separates ions Mass spectrum: presents information

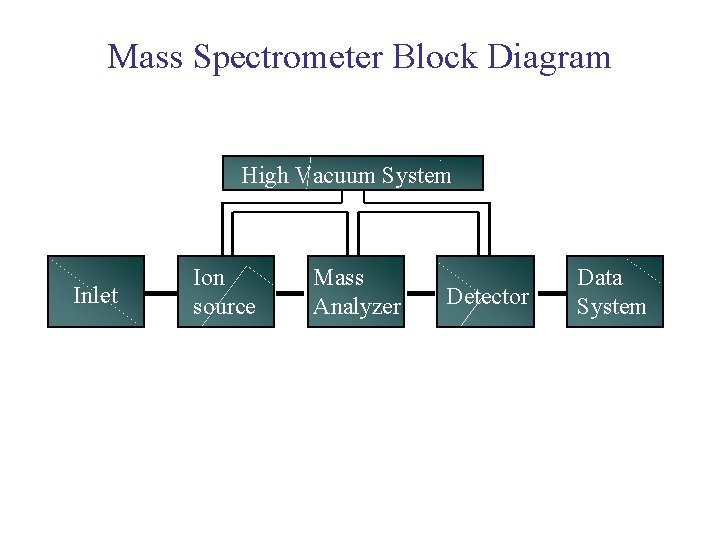
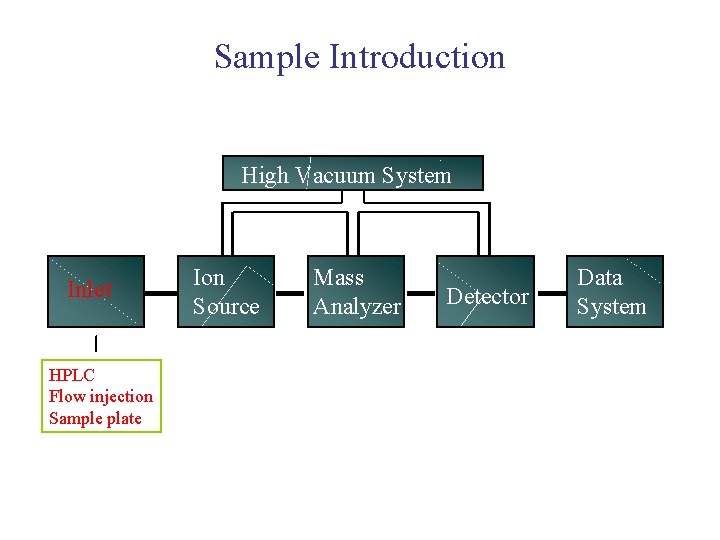
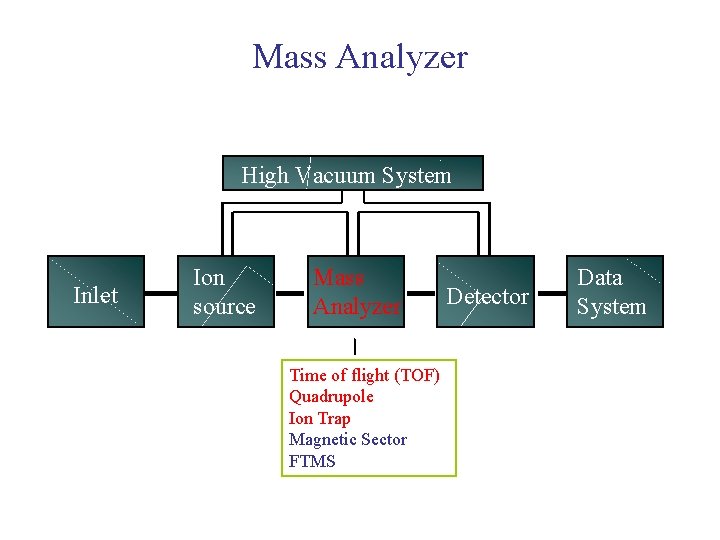
Mass Spectrometer Block Diagram High Vacuum System Inlet Ion source Mass Analyzer Detector Data System

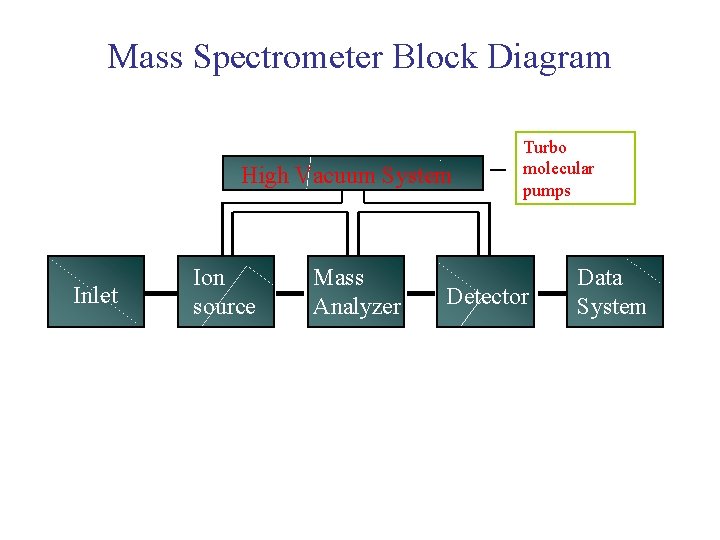
Mass Spectrometer Block Diagram High Vacuum System Inlet Ion source Mass Analyzer Turbo molecular pumps Detector Data System

Sample Introduction High Vacuum System Inlet HPLC Flow injection Sample plate Ion Source Mass Analyzer Detector Data System

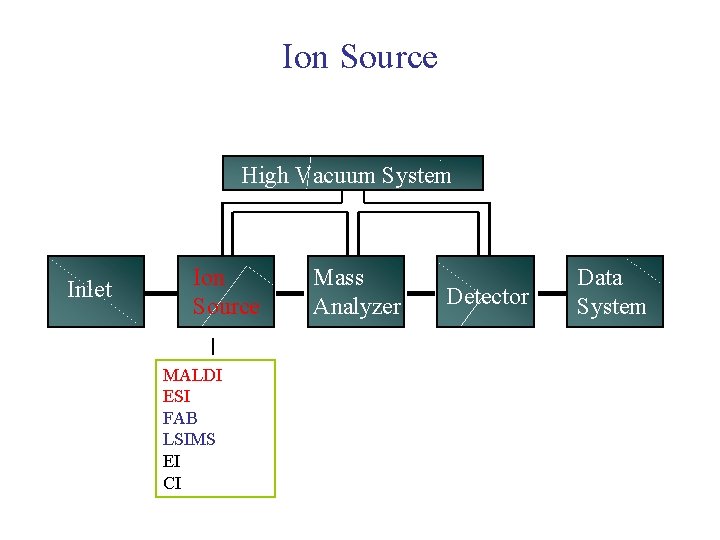
Ion Source High Vacuum System Inlet Ion Source MALDI ESI FAB LSIMS EI CI Mass Analyzer Detector Data System

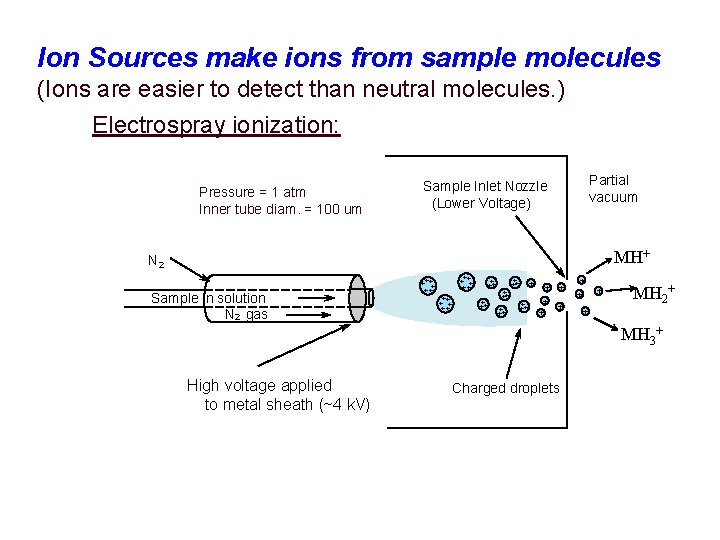
Ion Sources make ions from sample molecules (Ions are easier to detect than neutral molecules. ) Electrospray ionization: Pressure = 1 atm Inner tube diam. = 100 um Partial vacuum Sample Inlet Nozzle (Lower Voltage) MH+ N 2 Sample in solution N 2 gas + + ++ ++ ++ + ++ + ++ + + + + MH 2+ + MH 3+ High voltage applied to metal sheath (~4 k. V) Charged droplets

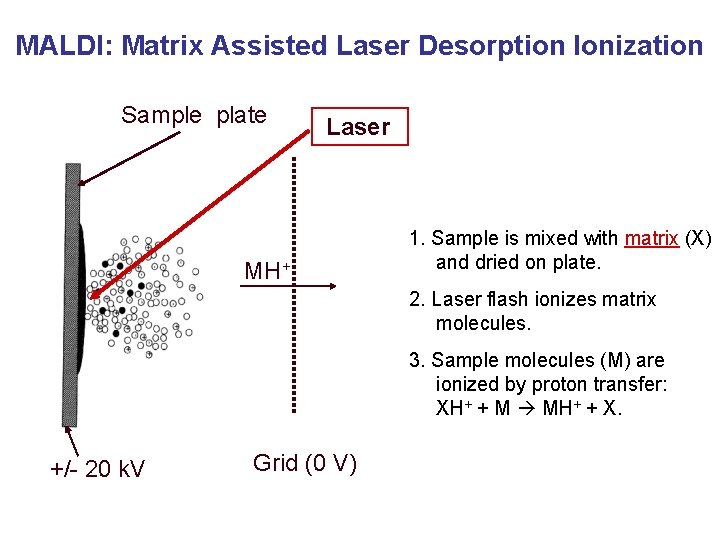
MALDI: Matrix Assisted Laser Desorption Ionization Sample plate Laser hn MH+ 1. Sample is mixed with matrix (X) and dried on plate. 2. Laser flash ionizes matrix molecules. 3. Sample molecules (M) are ionized by proton transfer: XH+ + M MH+ + X. +/- 20 k. V Grid (0 V)

Mass Analyzer High Vacuum System Inlet Ion source Mass Analyzer Time of flight (TOF) Quadrupole Ion Trap Magnetic Sector FTMS Detector Data System

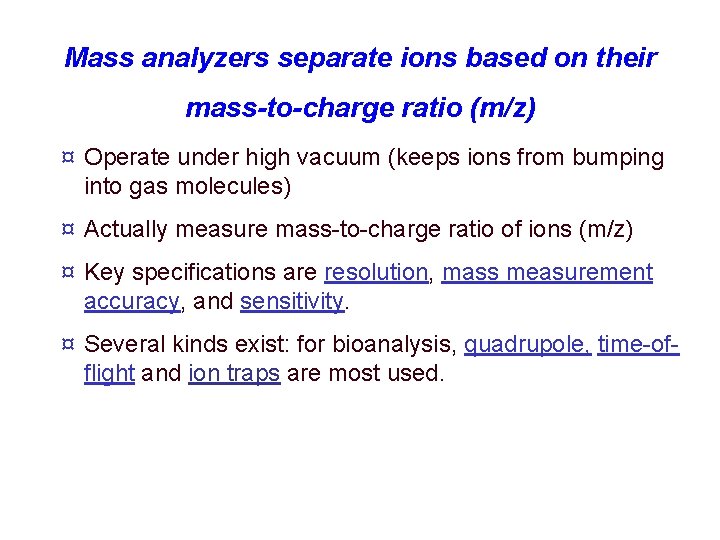
Mass analyzers separate ions based on their mass-to-charge ratio (m/z) ¤ Operate under high vacuum (keeps ions from bumping into gas molecules) ¤ Actually measure mass-to-charge ratio of ions (m/z) ¤ Key specifications are resolution, mass measurement accuracy, and sensitivity. ¤ Several kinds exist: for bioanalysis, quadrupole, time-offlight and ion traps are most used.

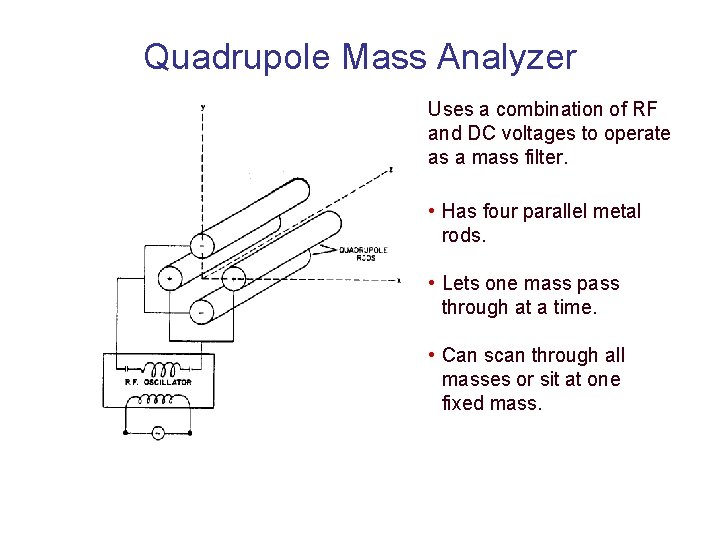
Quadrupole Mass Analyzer Uses a combination of RF and DC voltages to operate as a mass filter. • Has four parallel metal rods. • Lets one mass pass through at a time. • Can scan through all masses or sit at one fixed mass.

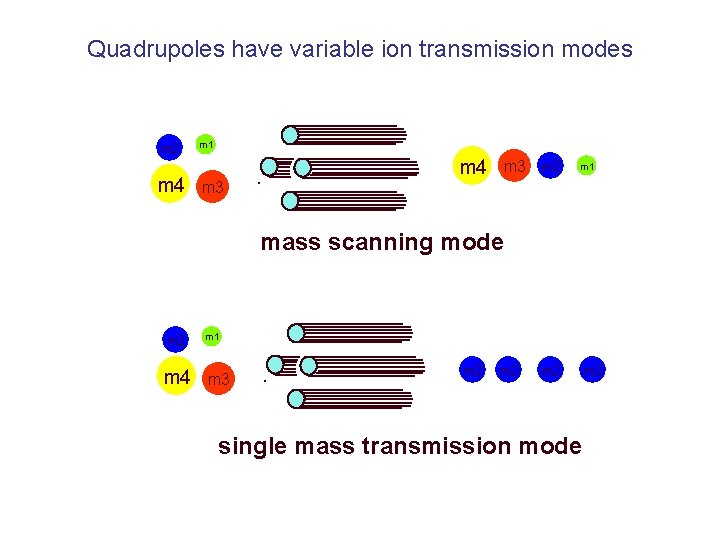
Quadrupoles have variable ion transmission modes m 2 m 4 m 1 m 4 m 3 m 2 m 1 m 3 mass scanning mode m 2 m 4 m 1 m 3 m 2 m 2 single mass transmission mode m 2

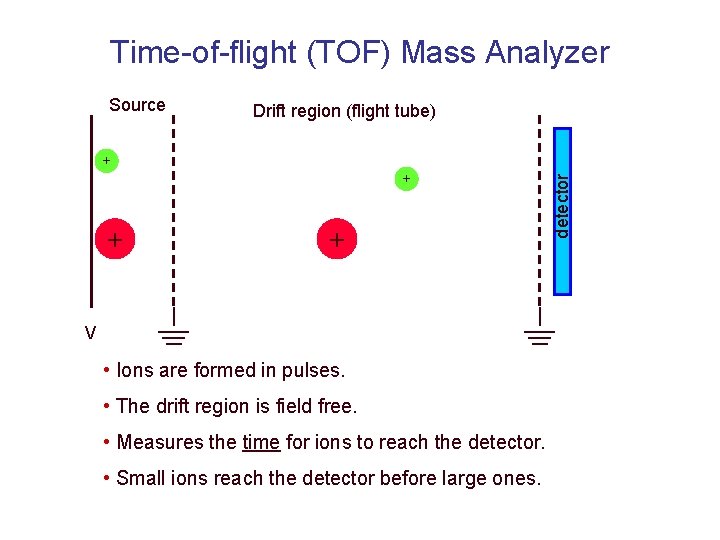
Time-of-flight (TOF) Mass Analyzer Drift region (flight tube) + + V • Ions are formed in pulses. • The drift region is field free. • Measures the time for ions to reach the detector. • Small ions reach the detector before large ones. detector Source

Ion Trap Mass Analyzer Top View Cut away side view

Detector High Vacuum System Inlet Ion source Mass Analyzer Detector Data System Microchannel Plate Electron Multiplier Hybrid with photomultiplier

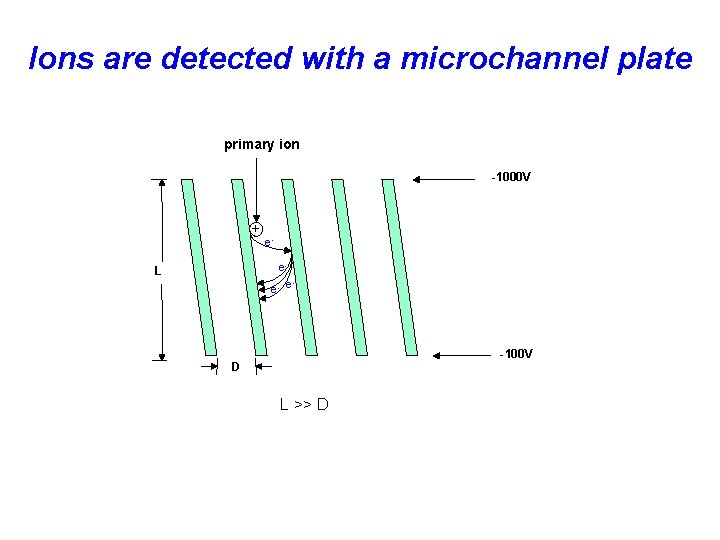
Ions are detected with a microchannel plate primary ion -1000 V + ee- L e- e -100 V D L >> D

Data System High Vacuum System Inlet Ion source Mass Analyzer Detector Data System PC Sun SPARK Station DEC Station

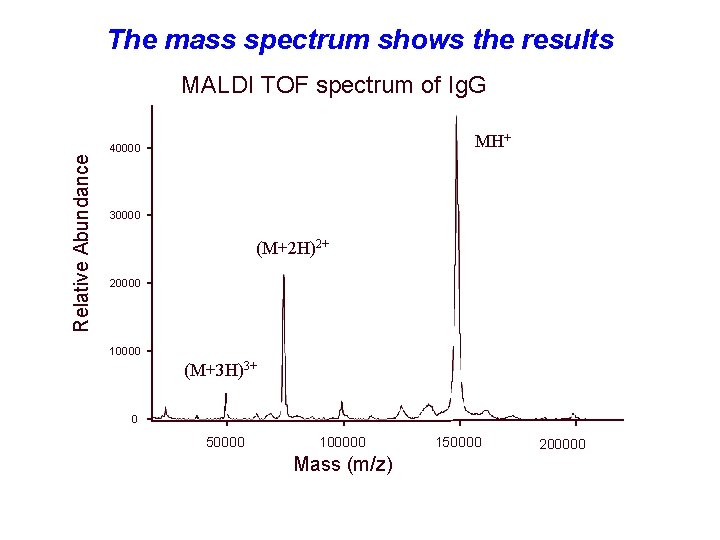
The mass spectrum shows the results Relative Abundance MALDI TOF spectrum of Ig. G MH+ 40000 30000 (M+2 H)2+ 20000 10000 (M+3 H)3+ 0 50000 100000 Mass (m/z) 150000 200000

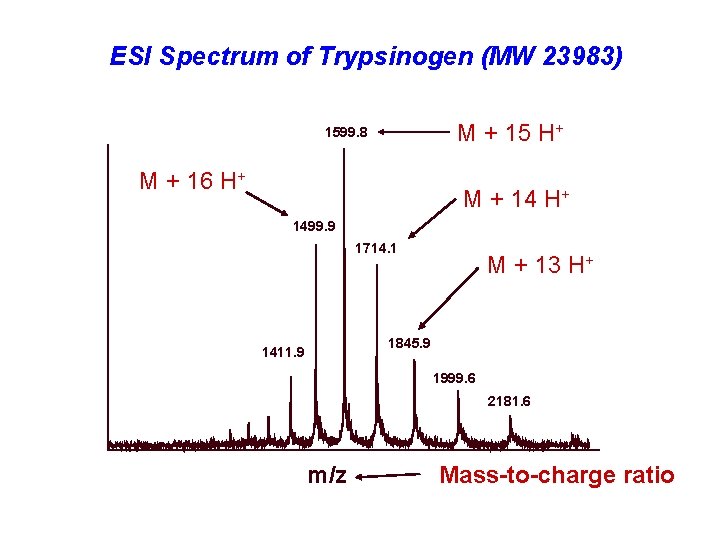
ESI Spectrum of Trypsinogen (MW 23983) M + 15 H+ 1599. 8 M + 16 H+ M + 14 H+ 1499. 9 1714. 1 M + 13 H+ 1845. 9 1411. 9 1999. 6 2181. 6 m/z Mass-to-charge ratio

How do mass spectrometers get their names? Types of ion sources: • Electrospray (ESI) • Matrix Assisted Laser Desorption Ionization (MALDI) Types of mass analyzers: • Quadrupole (Quad, Q) • Ion Trap • Time-of-Flight (TOF) - Either source type can work with either analyzer type: “MALDITOF, ” “ESI-Quad. ” - Analyzers can be combined to create “hybrid” instruments. ESIQQQ, MALDI QQ TOF, Q Trap

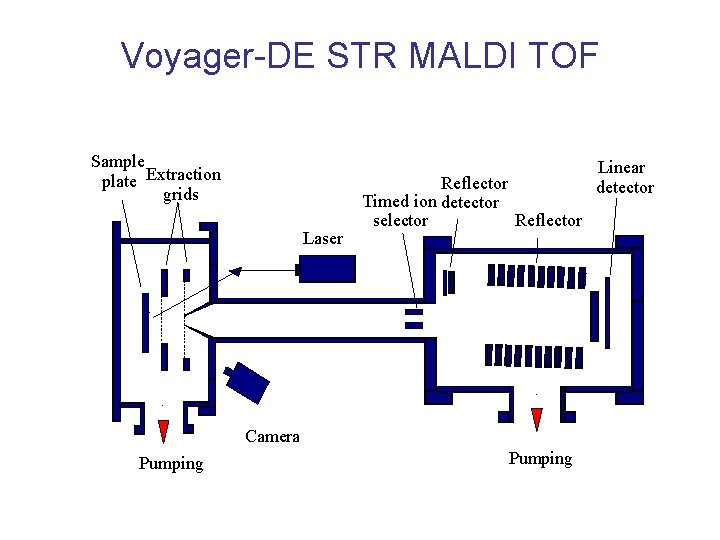
Voyager-DE STR MALDI TOF Sample plate Extraction grids Laser Reflector Timed ion detector selector Reflector Camera Pumping Linear detector

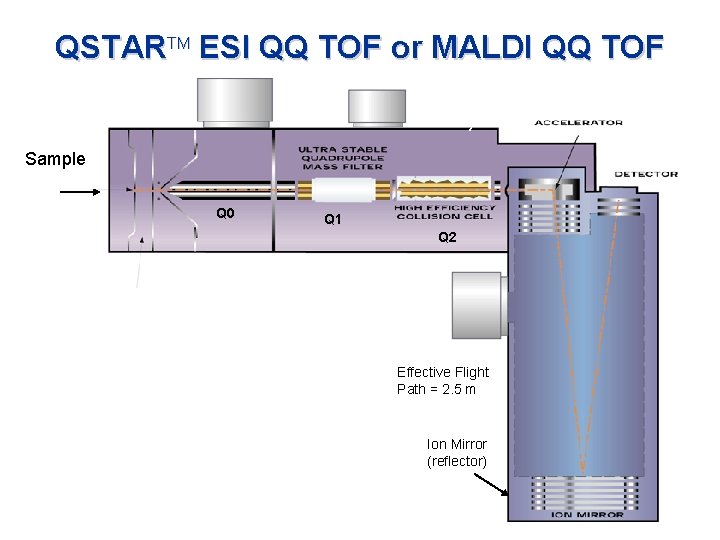
QSTARTM ESI QQ TOF or MALDI QQ TOF Sample Q 0 Q 1 Q 2 Effective Flight Path = 2. 5 m Ion Mirror (reflector)

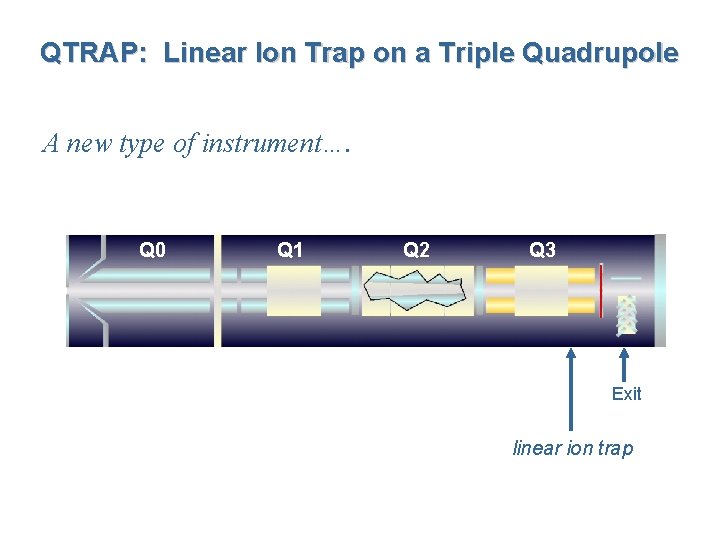
QTRAP: Linear Ion Trap on a Triple Quadrupole A new type of instrument…. Q 0 Q 1 Q 2 Q 3 Exit linear ion trap

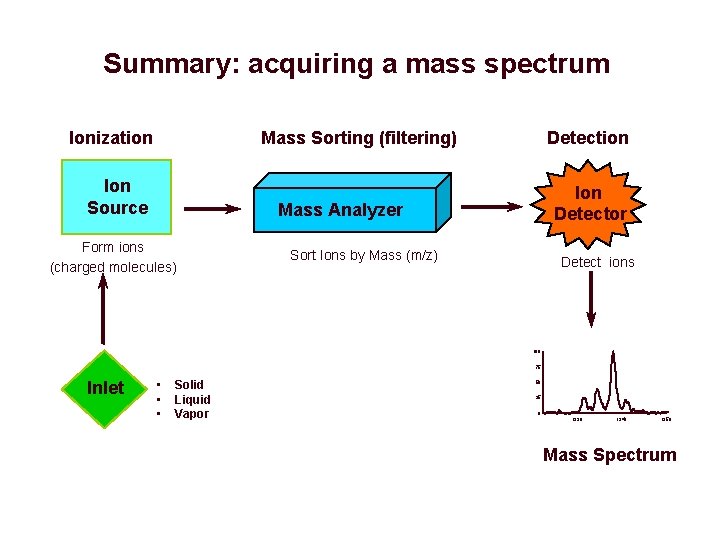
Summary: acquiring a mass spectrum Ionization Mass Sorting (filtering) Ion Source Detection Ion Detector Mass Analyzer Form ions (charged molecules) Sort Ions by Mass (m/z) Detect ions 100 75 Inlet • • • Solid Liquid Vapor 50 25 0 1330 1340 1350 Mass Spectrum

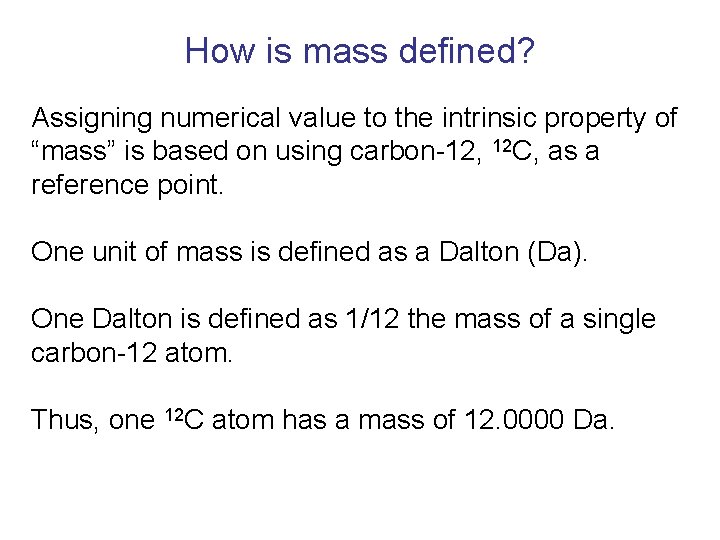
How is mass defined? Assigning numerical value to the intrinsic property of “mass” is based on using carbon-12, 12 C, as a reference point. One unit of mass is defined as a Dalton (Da). One Dalton is defined as 1/12 the mass of a single carbon-12 atom. Thus, one 12 C atom has a mass of 12. 0000 Da.

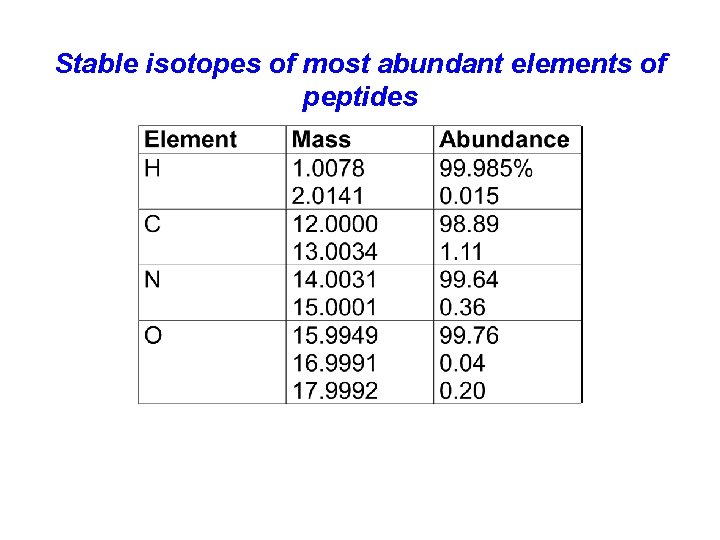
Isotopes +Most elements have more than one stable isotope. For example, most carbon atoms have a mass of 12 Da, but in nature, 1. 1% of C atoms have an extra neutron, making their mass 13 Da. +Why do we care? Mass spectrometers can “see” isotope peaks if their resolution is high enough. If an MS instrument has resolution high enough to resolve these isotopes, better mass accuracy is achieved.

Stable isotopes of most abundant elements of peptides

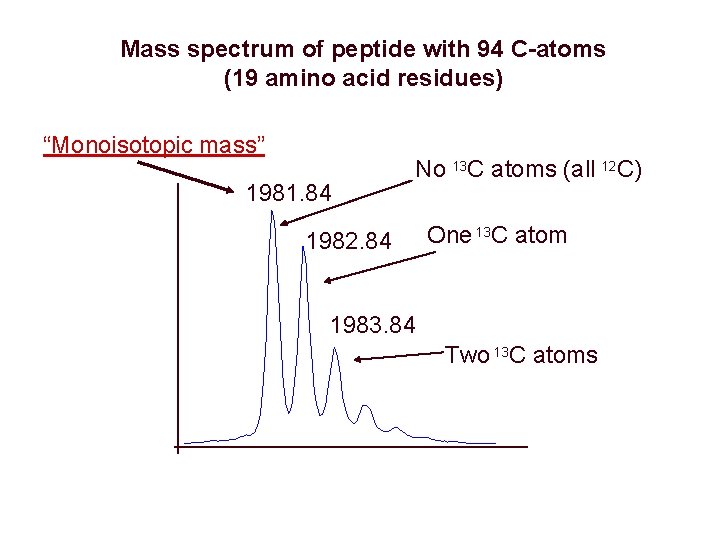
Mass spectrum of peptide with 94 C-atoms (19 amino acid residues) “Monoisotopic mass” 1981. 84 No 13 C atoms (all 12 C) 1982. 84 One 13 C atom 1983. 84 Two 13 C atoms

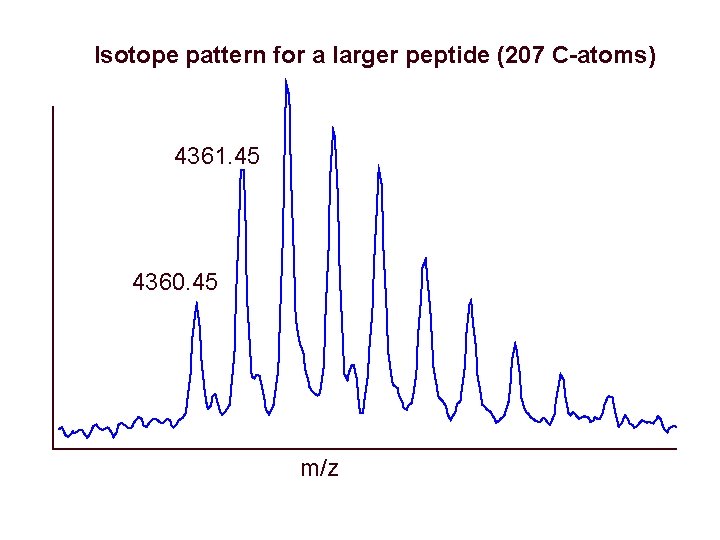
Isotope pattern for a larger peptide (207 C-atoms) 4361. 45 4360. 45 m/z

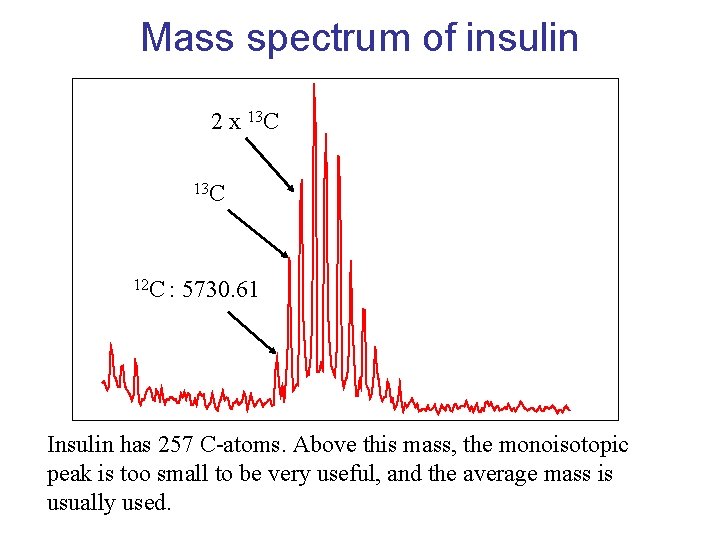
Mass spectrum of insulin 2 x 13 C 12 C : 5730. 61 Insulin has 257 C-atoms. Above this mass, the monoisotopic peak is too small to be very useful, and the average mass is usually used.

Monoisotopic mass When the isotopes are clearly resolved the monoisotopic mass is used as it is the most accurate measurement.

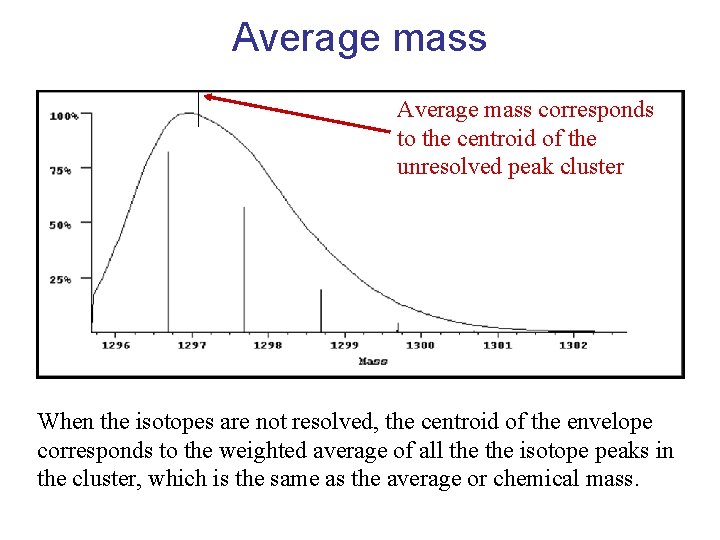
Average mass corresponds to the centroid of the unresolved peak cluster When the isotopes are not resolved, the centroid of the envelope corresponds to the weighted average of all the isotope peaks in the cluster, which is the same as the average or chemical mass.

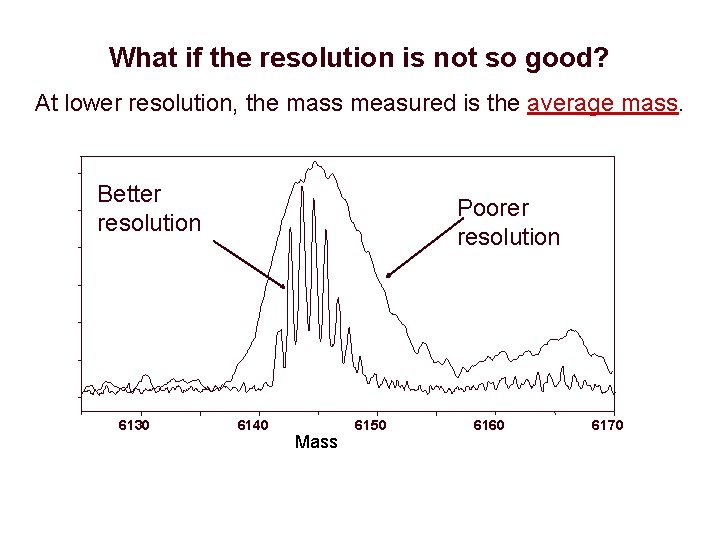
What if the resolution is not so good? At lower resolution, the mass measured is the average mass. Better resolution 6130 Poorer resolution 6140 Mass 6150 6160 6170

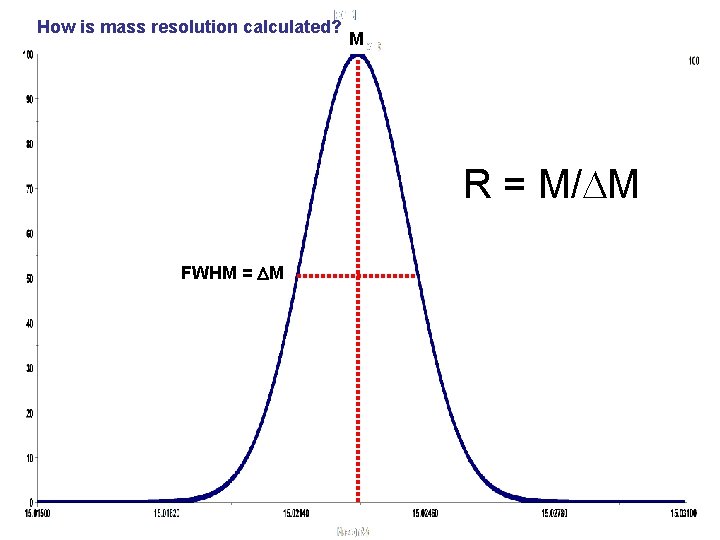
How is mass resolution calculated? M R = M/DM FWHM = DM

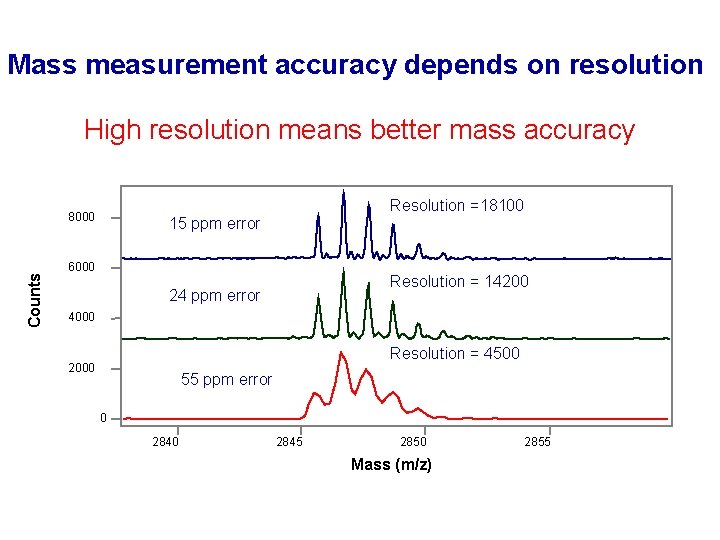
Mass measurement accuracy depends on resolution High resolution means better mass accuracy Counts 8000 Resolution =18100 15 ppm error 6000 Resolution = 14200 24 ppm error 4000 Resolution = 4500 2000 55 ppm error 0 2845 2850 Mass (m/z) 2855

How do we achieve superior mass resolution? Reflector TOF Mass Analyzer Delayed Extraction on a MALDI source

Important performance factors Mass accuracy: How accurate is the mass measurement? Resolution: How well separated are the peaks from each other? Sensitivity: How small an amount can be analyzed?

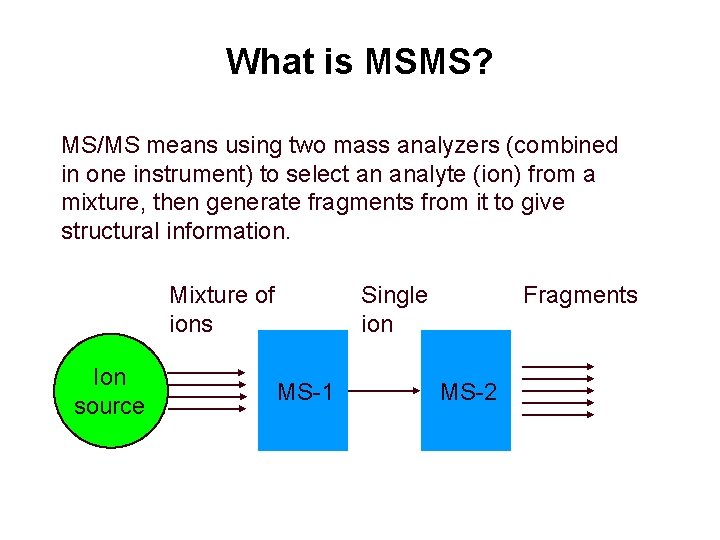
What is MSMS? MS/MS means using two mass analyzers (combined in one instrument) to select an analyte (ion) from a mixture, then generate fragments from it to give structural information. Mixture of ions Ion source Single ion MS-1 Fragments MS-2

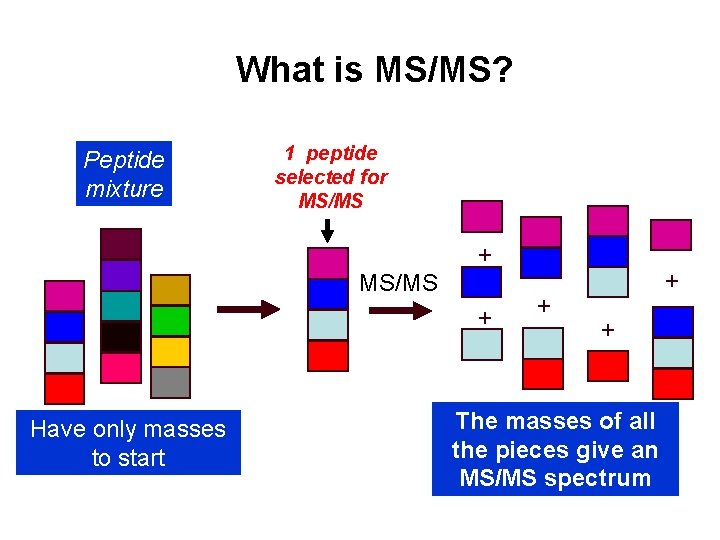
What is MS/MS? Peptide mixture 1 peptide selected for MS/MS + Have only masses to start + + + The masses of all the pieces give an MS/MS spectrum

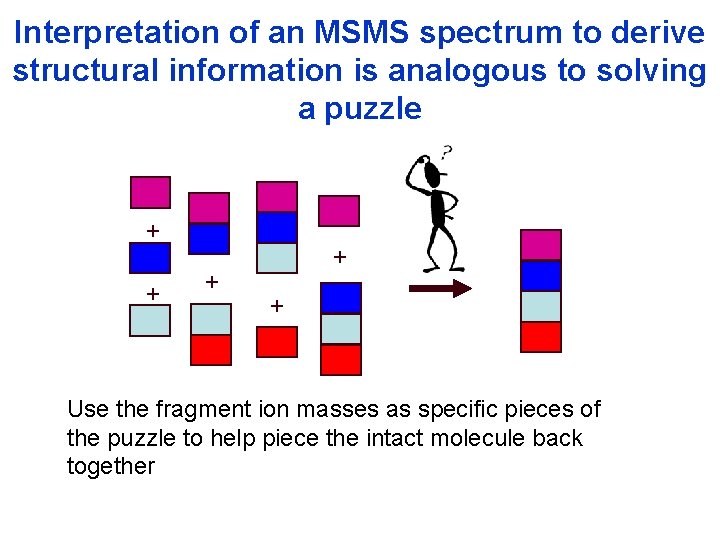
Interpretation of an MSMS spectrum to derive structural information is analogous to solving a puzzle + + + Use the fragment ion masses as specific pieces of the puzzle to help piece the intact molecule back together

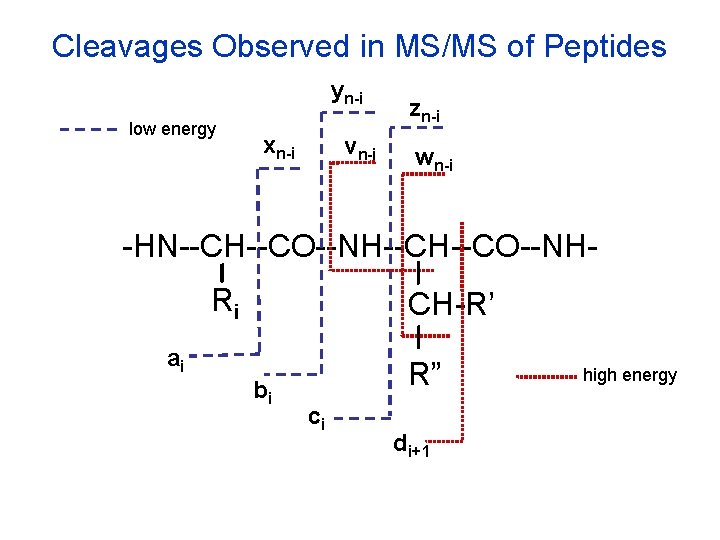
Cleavages Observed in MS/MS of Peptides yn-i low energy xn-i vn-i zn-i wn-i -HN--CH--CO--NHRi CH-R’ ai bi R” ci di+1 high energy

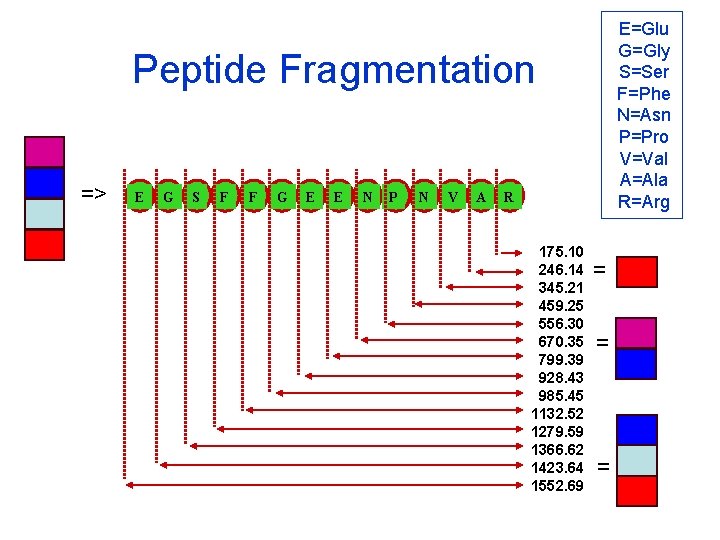
E=Glu G=Gly S=Ser F=Phe N=Asn P=Pro V=Val A=Ala R=Arg Peptide Fragmentation => E G S F F G E E N P N V A R 175. 10 246. 14 345. 21 459. 25 556. 30 670. 35 799. 39 928. 43 985. 45 1132. 52 1279. 59 1366. 62 1423. 64 1552. 69 = = =

Protein Identification 1. Peptide Mass Finger Printing (PMF) from MS data 2. Database search using fragment ion masses from MS/MS data 3. Sequence Tags from MS/MS data

PROBLEM Bank President Biologist Who robbed the bank? What protein was isolated?

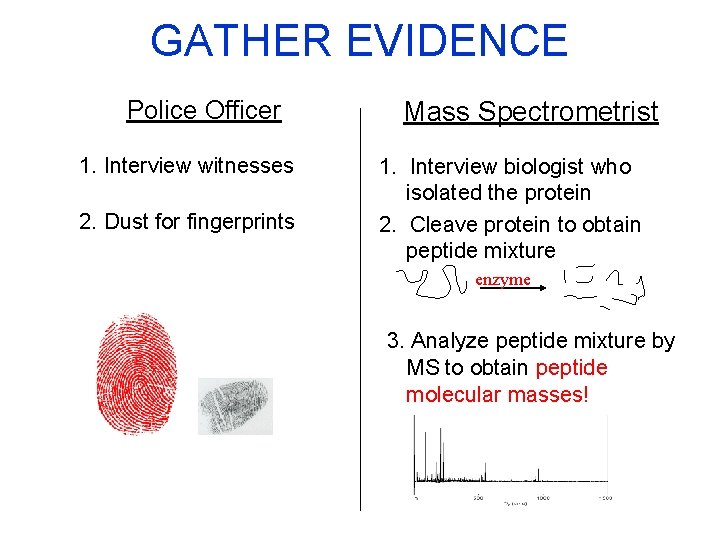
GATHER EVIDENCE Police Officer 1. Interview witnesses 2. Dust for fingerprints Mass Spectrometrist 1. Interview biologist who isolated the protein 2. Cleave protein to obtain peptide mixture enzyme 3. Analyze peptide mixture by MS to obtain peptide molecular masses!

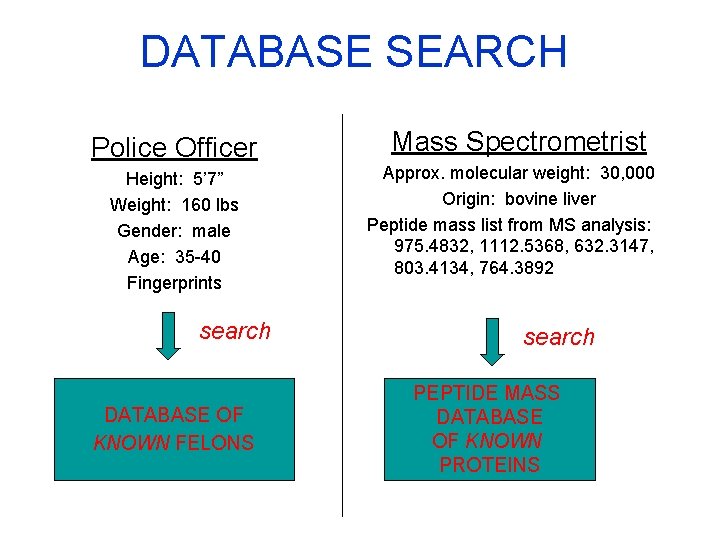
DATABASE SEARCH Police Officer Height: 5’ 7” Weight: 160 lbs Gender: male Age: 35 -40 Fingerprints search DATABASE OF KNOWN FELONS Mass Spectrometrist Approx. molecular weight: 30, 000 Origin: bovine liver Peptide mass list from MS analysis: 975. 4832, 1112. 5368, 632. 3147, 803. 4134, 764. 3892 search PEPTIDE MASS DATABASE OF KNOWN PROTEINS

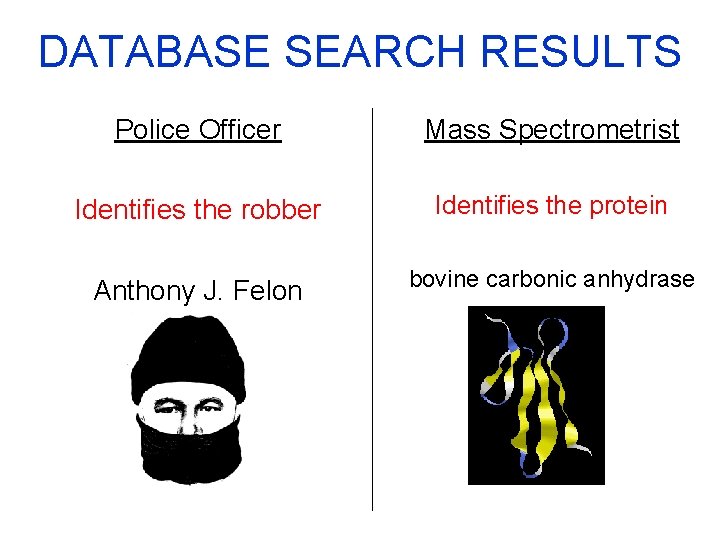
DATABASE SEARCH RESULTS Police Officer Mass Spectrometrist Identifies the robber Identifies the protein Anthony J. Felon bovine carbonic anhydrase

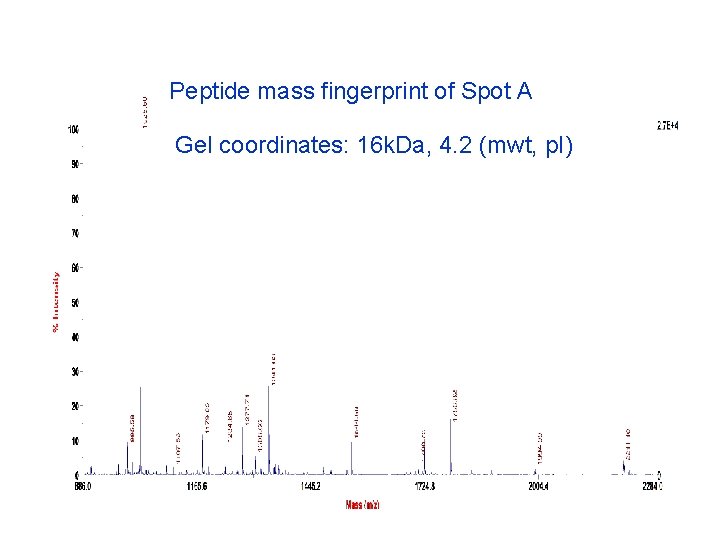
Peptide mass fingerprint of Spot A Gel coordinates: 16 k. Da, 4. 2 (mwt, p. I)

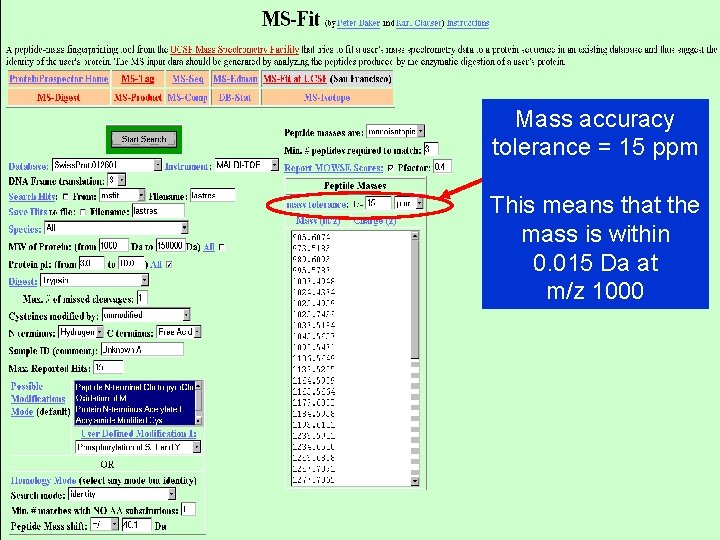
Mass accuracy tolerance = 15 ppm This means that the mass is within 0. 015 Da at m/z 1000



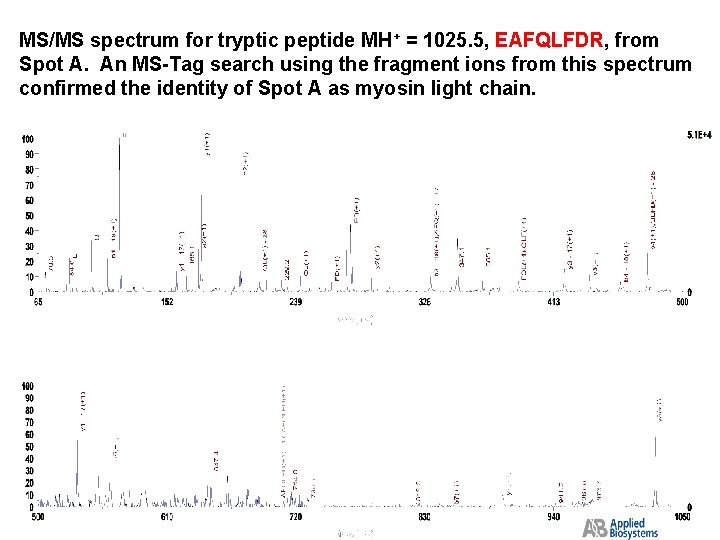
MS/MS spectrum for tryptic peptide MH+ = 1025. 5, EAFQLFDR, from Spot A. An MS-Tag search using the fragment ions from this spectrum confirmed the identity of Spot A as myosin light chain.

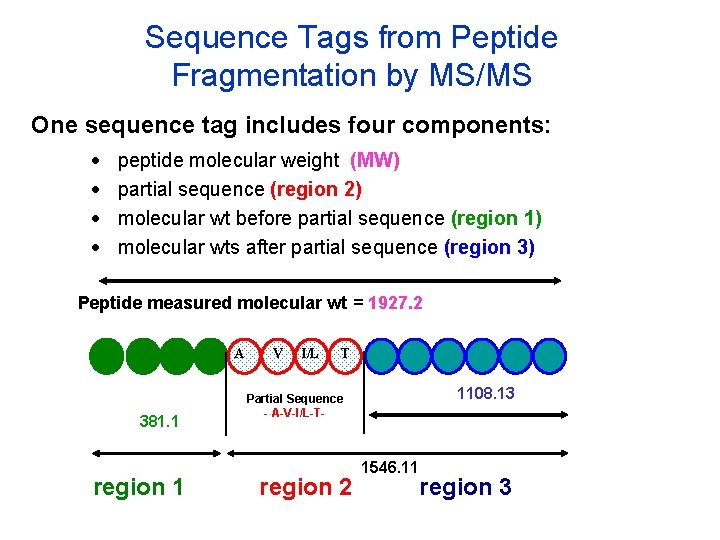
Sequence Tags from Peptide Fragmentation by MS/MS One sequence tag includes four components: · · peptide molecular weight (MW) partial sequence (region 2) molecular wt before partial sequence (region 1) molecular wts after partial sequence (region 3) Peptide measured molecular wt = 1927. 2 A 381. 1 region 1 V I/L T 1108. 13 Partial Sequence - A-V-I/L-T- region 2 1546. 11 region 3

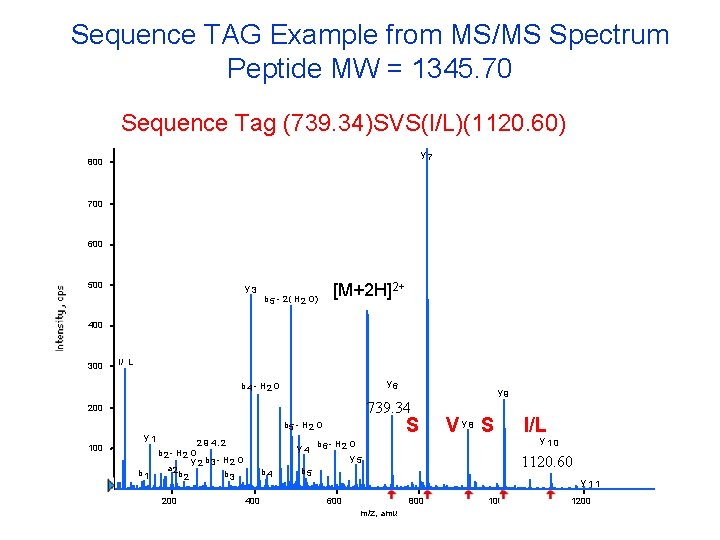
Sequence TAG Example from MS/MS Spectrum Peptide MW = 1345. 70 Sequence Tag (739. 34)SVS(I/L)(1120. 60) y 7 800 700 600 500 y 3 b 5 - 2 ( H 2 O) [M+2 H]2+ 400 300 I/ L y 6 b 4 - H 2 O 739. 34 200 100 S b 5 - H 2 O y 1 2 9 4. 2 b 2 - H 2 O y 2 b 3 - H 2 O a 2 b 1 b 2 b 3 200 b 4 400 y 9 Vy 8 S y 4 b 6 - H 2 O y 5 b 5 600 y 10 1120. 60 y 11 800 m/z, amu I/L 1000 1200

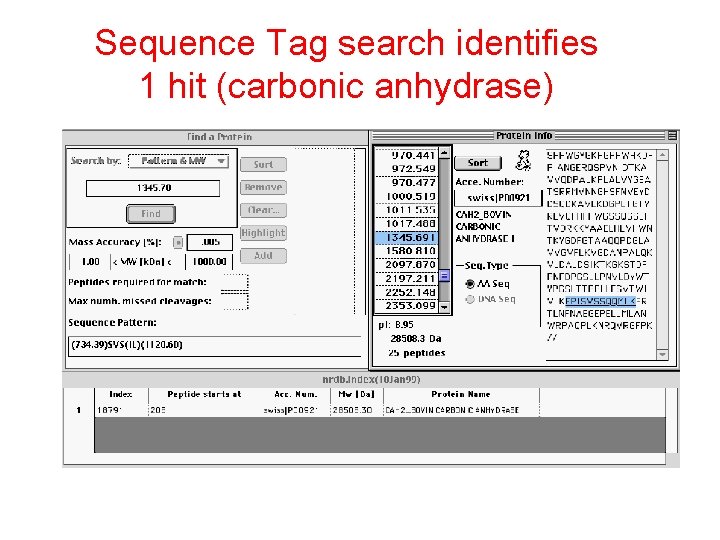
Sequence Tag search identifies 1 hit (carbonic anhydrase)

Acknowledgements We thank the Applied Biosystems Mass Spectrometry Applications Laboratory for allowing the use of some of their slides for this presentation.