MAP estimation in MRFs via rank aggregation Rahul
![MAP via rank-aggregation n [Step 1] Decompose graph potentials into a convex combination of MAP via rank-aggregation n [Step 1] Decompose graph potentials into a convex combination of](https://slidetodoc.com/presentation_image/5d479ec825ab37c4987303ff8f9edaa5/image-4.jpg)
![Rank aggregation (contd. ) n [Step 2] Perform top-k MAP estimation on each constituent Rank aggregation (contd. ) n [Step 2] Perform top-k MAP estimation on each constituent](https://slidetodoc.com/presentation_image/5d479ec825ab37c4987303ff8f9edaa5/image-5.jpg)
![Rank-Merge n [Step 3] Merge the ranked lists using the aggregate function Score(x) = Rank-Merge n [Step 3] Merge the ranked lists using the aggregate function Score(x) =](https://slidetodoc.com/presentation_image/5d479ec825ab37c4987303ff8f9edaa5/image-6.jpg)

- Slides: 15

MAP estimation in MRFs via rank aggregation Rahul Gupta Sunita Sarawagi (IBM India Research Lab) (IIT Bombay)

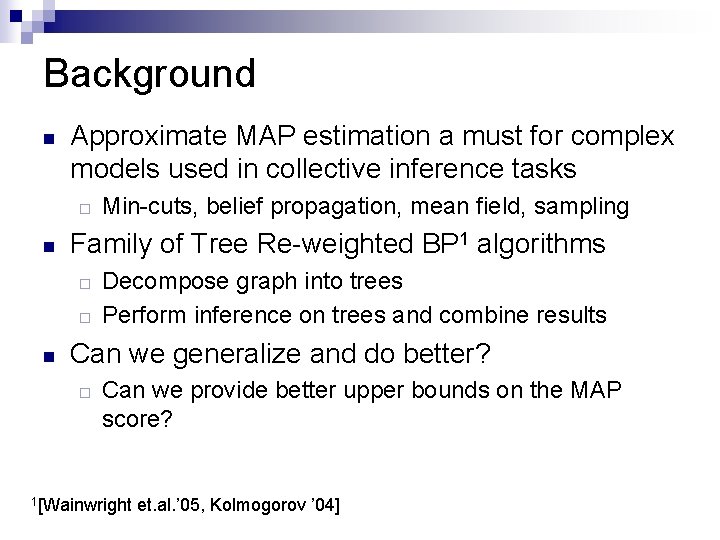
Background n Approximate MAP estimation a must for complex models used in collective inference tasks ¨ n Family of Tree Re-weighted BP 1 algorithms ¨ ¨ n Min-cuts, belief propagation, mean field, sampling Decompose graph into trees Perform inference on trees and combine results Can we generalize and do better? ¨ Can we provide better upper bounds on the MAP score? 1[Wainwright et. al. ’ 05, Kolmogorov ’ 04]

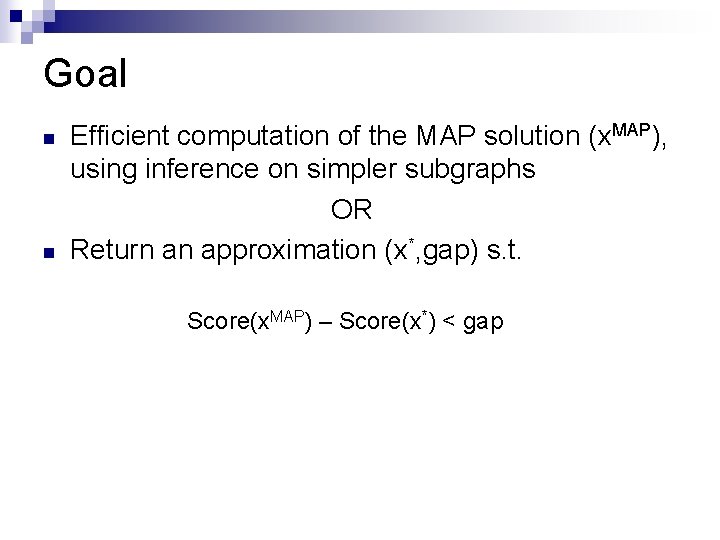
Goal n n Efficient computation of the MAP solution (x. MAP), using inference on simpler subgraphs OR Return an approximation (x*, gap) s. t. Score(x. MAP) – Score(x*) < gap
![MAP via rankaggregation n Step 1 Decompose graph potentials into a convex combination of MAP via rank-aggregation n [Step 1] Decompose graph potentials into a convex combination of](https://slidetodoc.com/presentation_image/5d479ec825ab37c4987303ff8f9edaa5/image-4.jpg)
MAP via rank-aggregation n [Step 1] Decompose graph potentials into a convex combination of simpler potentials ¨ E. g. Set of spanning trees that cover all edges = G => Score(x) + + = T 1 + T 2 + T 3 = Score 1(x) + Score 2(x) + Score 3(x)
![Rank aggregation contd n Step 2 Perform topk MAP estimation on each constituent Rank aggregation (contd. ) n [Step 2] Perform top-k MAP estimation on each constituent](https://slidetodoc.com/presentation_image/5d479ec825ab37c4987303ff8f9edaa5/image-5.jpg)
Rank aggregation (contd. ) n [Step 2] Perform top-k MAP estimation on each constituent and compute upper bound (ub) S 1 : x 1 x 2 x 3 x 4 x 5 x 6 x 7 x 8 ub = score 1(x 8) Si : x 7 x 20 x 4 x 1 x 9 x 11 x 8 x 2 + scorei(x 2) SL : x 15 x 6 x 2 x 8 x 22 x 3 x 5 x 4 + score. L(x 4) ties
![RankMerge n Step 3 Merge the ranked lists using the aggregate function Scorex Rank-Merge n [Step 3] Merge the ranked lists using the aggregate function Score(x) =](https://slidetodoc.com/presentation_image/5d479ec825ab37c4987303ff8f9edaa5/image-6.jpg)
Rank-Merge n [Step 3] Merge the ranked lists using the aggregate function Score(x) = i Scorei(x) Computed directly from the model x 2 x 1 x 8 x* (MAP estimate) x 1 x 2 x 3 x 4 x 5 x 6 x 7 x 8 x 7 x 20 x 4 x 1 x 9 x 11 x 8 x 2 x 15 x 6 x 2 x 8 x 22 x 3 x 5 x 4

Rank-Merge (contd. ) n If Score(x*) ≥ ub, then x. MAP = x* ¨ n If Score(x*) < ub, then Score(x. MAP) < ub ¨ ¨ n From the property of Rank. Merge algorithm From convexity of ‘max’ and decomposition of G Tighter bounds can be obtained by increasing k Can do even better ¨ Generate top-K and upper bounds ubi incrementally.

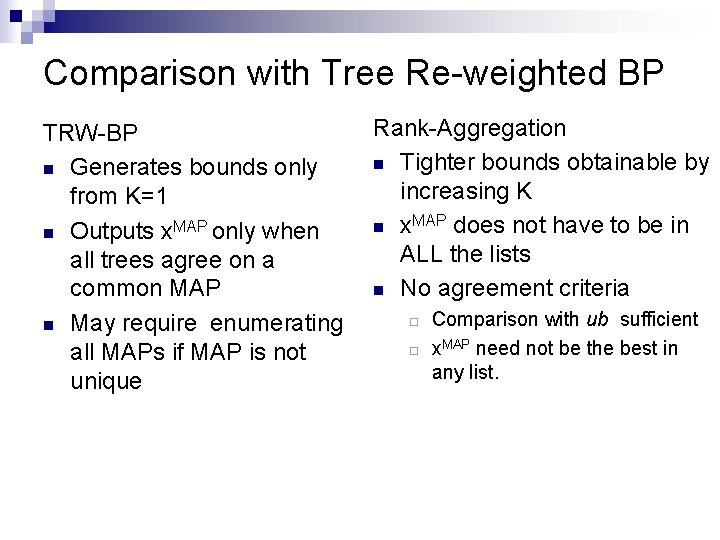
Comparison with Tree Re-weighted BP TRW-BP n Generates bounds only from K=1 n Outputs x. MAP only when all trees agree on a common MAP n May require enumerating all MAPs if MAP is not unique Rank-Aggregation n Tighter bounds obtainable by increasing K n x. MAP does not have to be in ALL the lists n No agreement criteria ¨ ¨ Comparison with ub sufficient x. MAP need not be the best in any list.

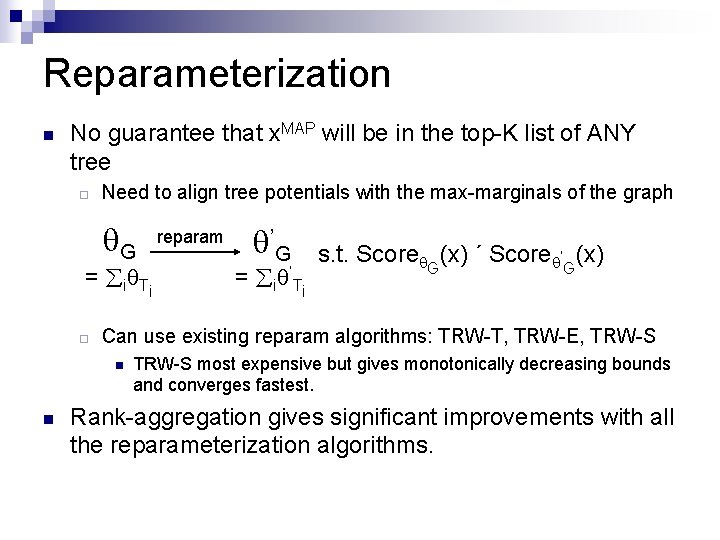
Reparameterization n No guarantee that x. MAP will be in the top-K list of ANY tree ¨ Need to align tree potentials with the max-marginals of the graph G = i T i ¨ ’G = i ’T i s. t. Score G(x) ´ Score ’G(x) Can use existing reparam algorithms: TRW-T, TRW-E, TRW-S n n reparam TRW-S most expensive but gives monotonically decreasing bounds and converges fastest. Rank-aggregation gives significant improvements with all the reparameterization algorithms.

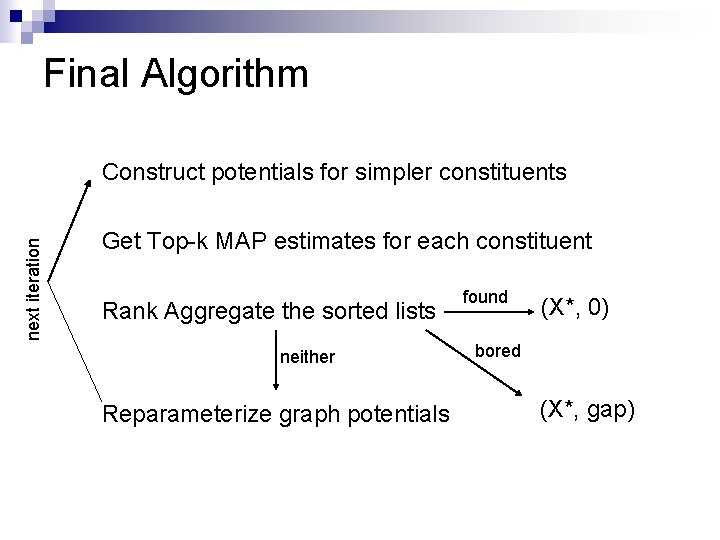
Final Algorithm next iteration Construct potentials for simpler constituents Get Top-k MAP estimates for each constituent Rank Aggregate the sorted lists neither Reparameterize graph potentials found (X*, 0) bored (X*, gap)

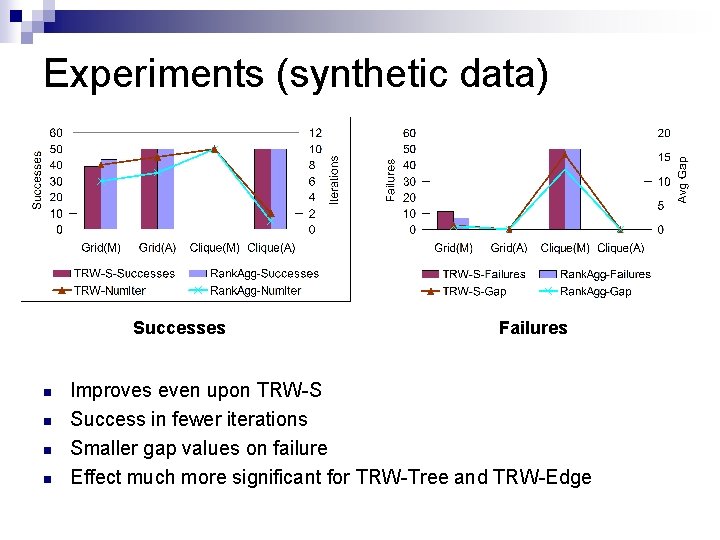
Experiments (synthetic data) Successes n n Failures Improves even upon TRW-S Success in fewer iterations Smaller gap values on failure Effect much more significant for TRW-Tree and TRW-Edge

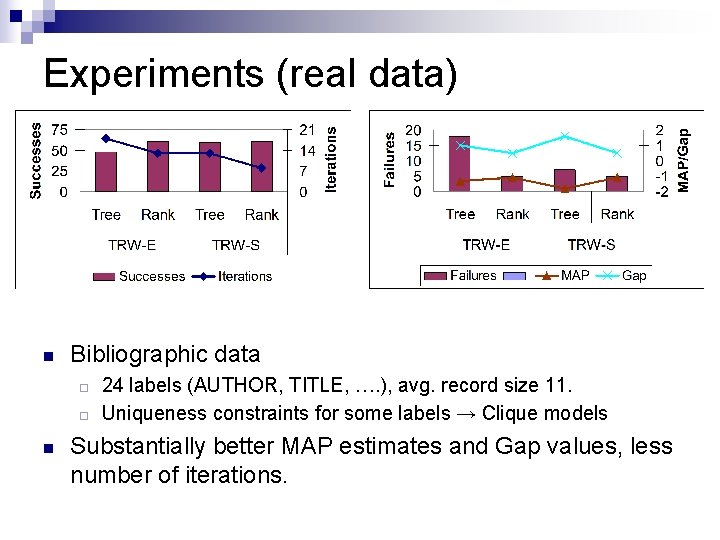
Experiments (real data) n Bibliographic data ¨ ¨ n 24 labels (AUTHOR, TITLE, …. ), avg. record size 11. Uniqueness constraints for some labels → Clique models Substantially better MAP estimates and Gap values, less number of iterations.

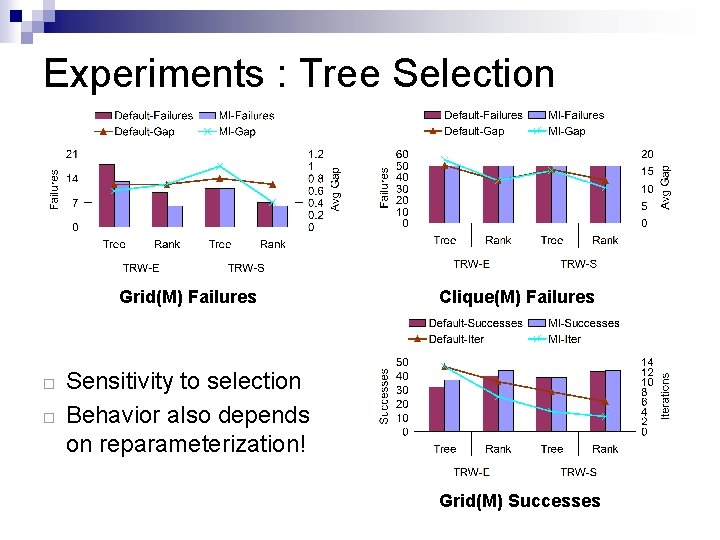
Experiments : Tree Selection Grid(M) Failures ¨ ¨ Clique(M) Failures Sensitivity to selection Behavior also depends on reparameterization! Grid(M) Successes

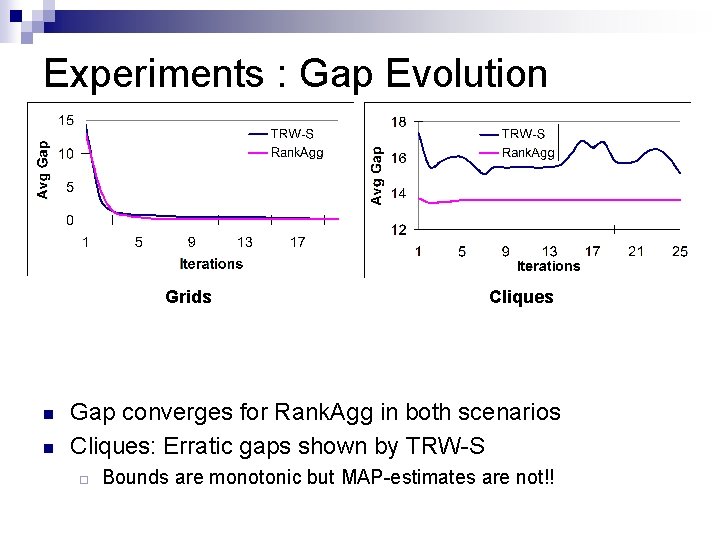
Experiments : Gap Evolution Grids n n Cliques Gap converges for Rank. Agg in both scenarios Cliques: Erratic gaps shown by TRW-S ¨ Bounds are monotonic but MAP-estimates are not!!

Summary and future work n Rank aggregation significantly better in MAPestimation ¨ ¨ ¨ n Fewer iterations, Much tighter bounds, Low k. No dependence on the tree-agreement criteria Can generalize to non-tree constituents as long as top -k is supported. Future work ¨ Collective inference on constrained models n ¨ Intelligent constraint generation Decrease sensitivity to tree selection