mandatory to put some order in such a
















- Slides: 55

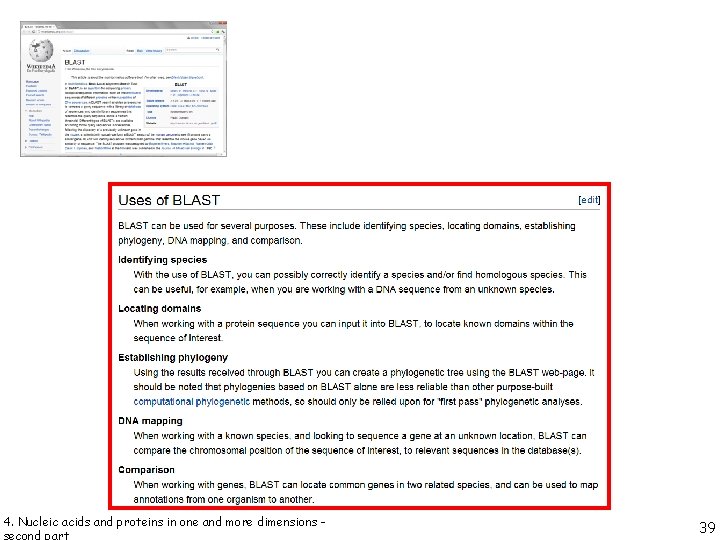
mandatory to put some order in such a vast wealth of structural knowledge 4. Nucleic acids and proteins in one and more dimensions - second part 1

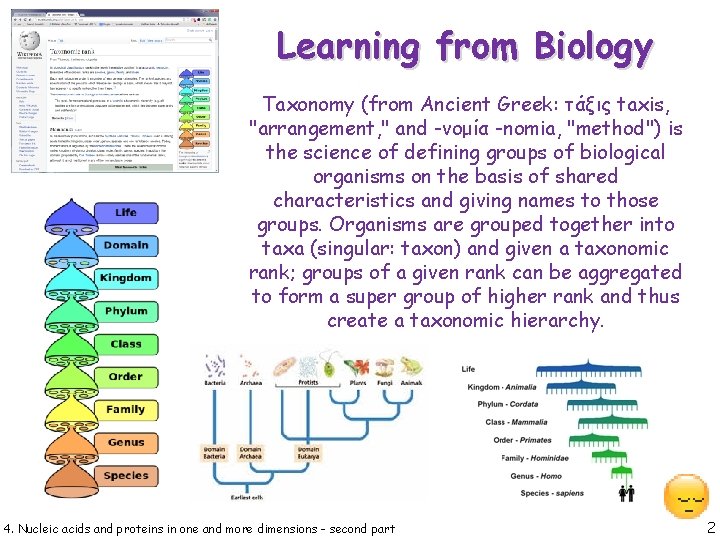
Learning from Biology Taxonomy (from Ancient Greek: τάξις taxis, "arrangement, " and -νομία -nomia, "method") is the science of defining groups of biological organisms on the basis of shared characteristics and giving names to those groups. Organisms are grouped together into taxa (singular: taxon) and given a taxonomic rank; groups of a given rank can be aggregated to form a super group of higher rank and thus create a taxonomic hierarchy. 4. Nucleic acids and proteins in one and more dimensions - second part 2

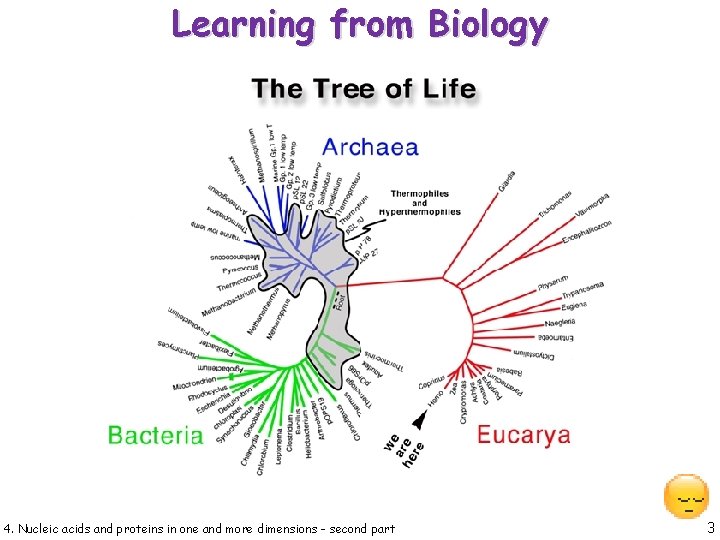
Learning from Biology 4. Nucleic acids and proteins in one and more dimensions - second part 3

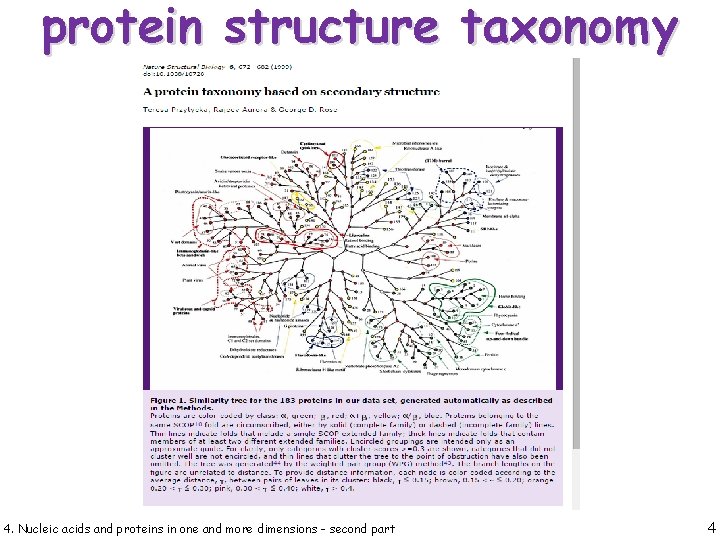
protein structure taxonomy 4. Nucleic acids and proteins in one and more dimensions - second part 4

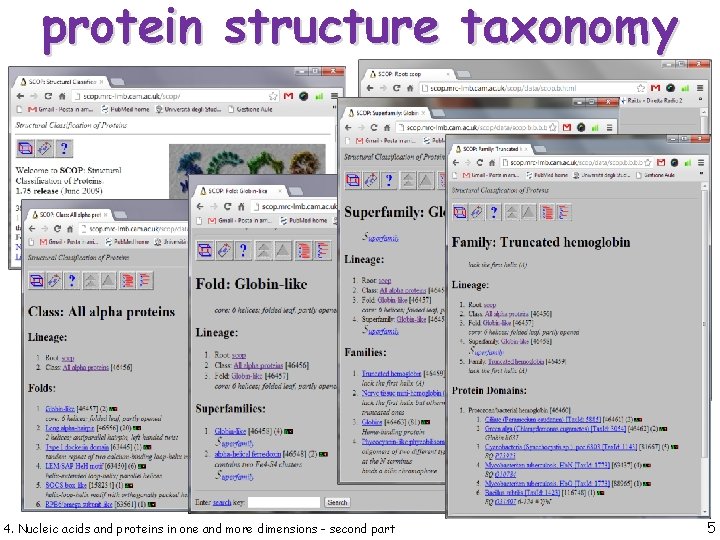
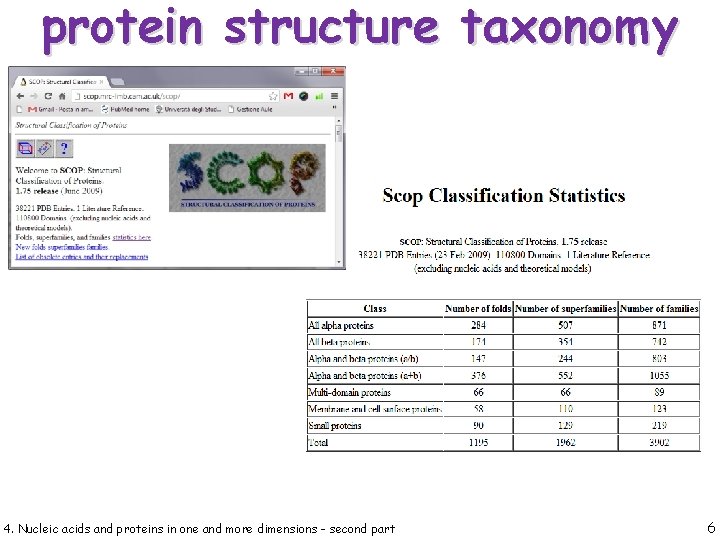
protein structure taxonomy 4. Nucleic acids and proteins in one and more dimensions - second part 5

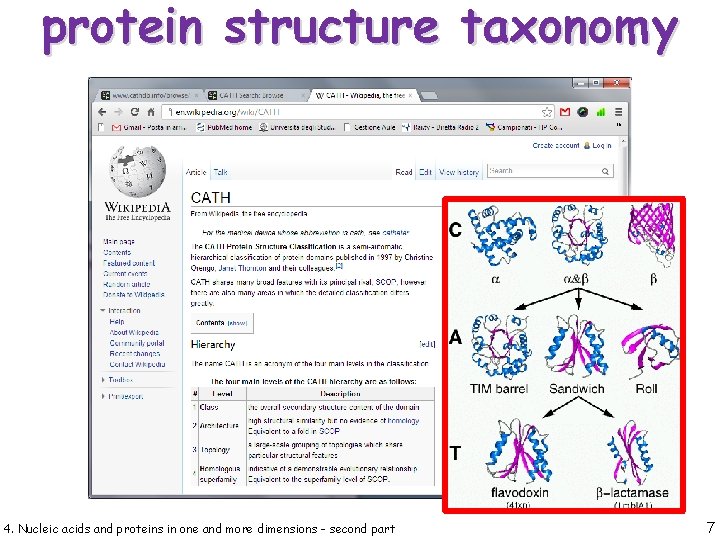
protein structure taxonomy 4. Nucleic acids and proteins in one and more dimensions - second part 6

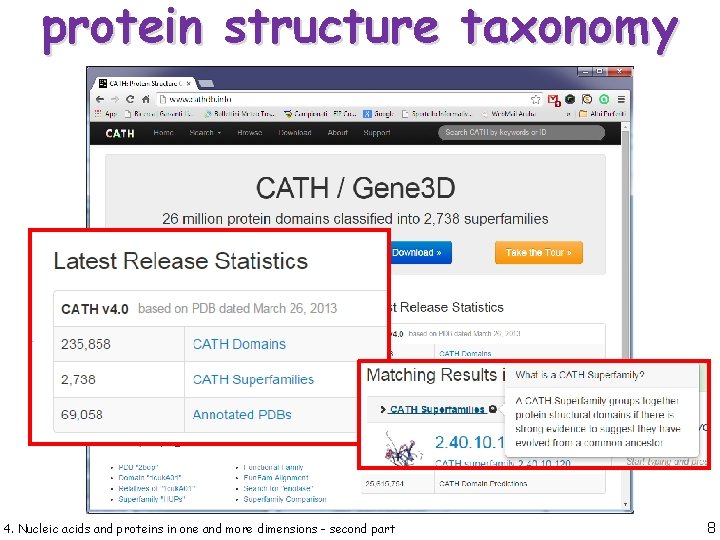
protein structure taxonomy 4. Nucleic acids and proteins in one and more dimensions - second part 7

protein structure taxonomy 4. Nucleic acids and proteins in one and more dimensions - second part 8

protein structure taxonomy 4. Nucleic acids and proteins in one and more dimensions - second part 9

protein structure taxonomy 4. Nucleic acids and proteins in one and more dimensions - second part 10

protein structure taxonomy red: mainly α green: mainly β yellow: αβ blue: low content of secondary structures 4. Nucleic acids and proteins in one and more dimensions - second part 11

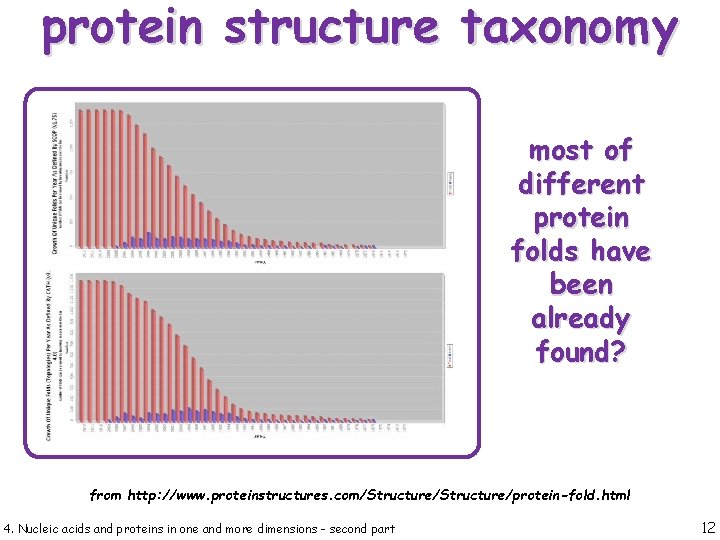
protein structure taxonomy most of different protein folds have been already found? from http: //www. proteinstructures. com/Structure/protein-fold. html 4. Nucleic acids and proteins in one and more dimensions - second part 12

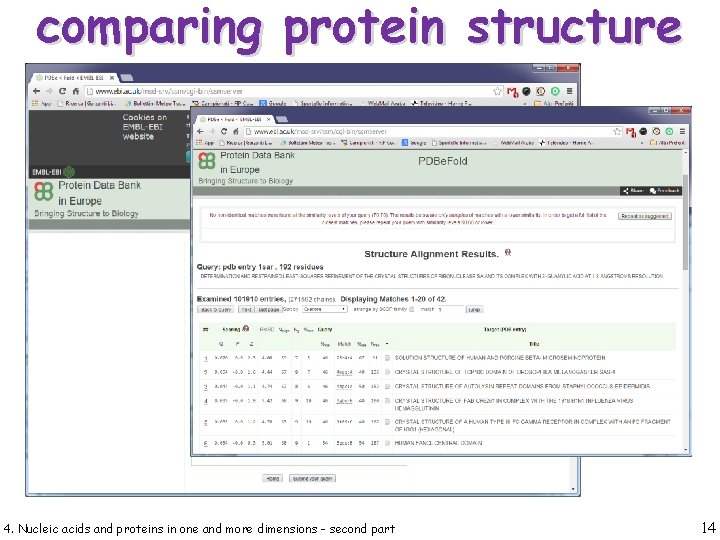
comparing protein structure 4. Nucleic acids and proteins in one and more dimensions - second part 13

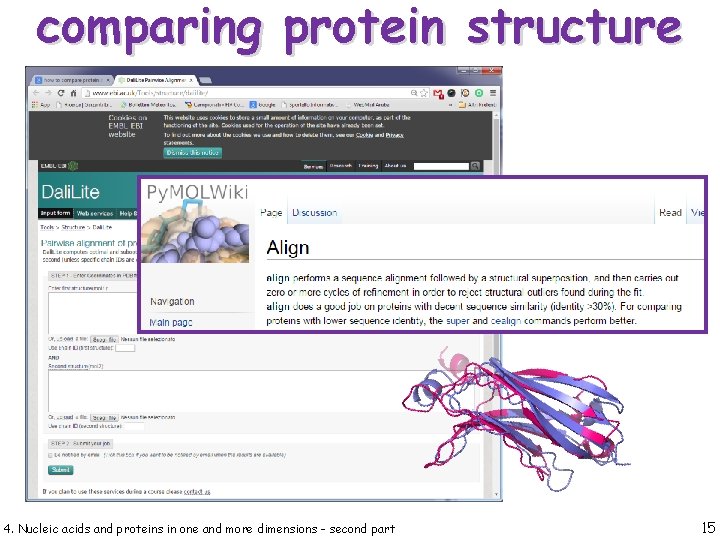
comparing protein structure 4. Nucleic acids and proteins in one and more dimensions - second part 14

comparing protein structure 4. Nucleic acids and proteins in one and more dimensions - second part 15

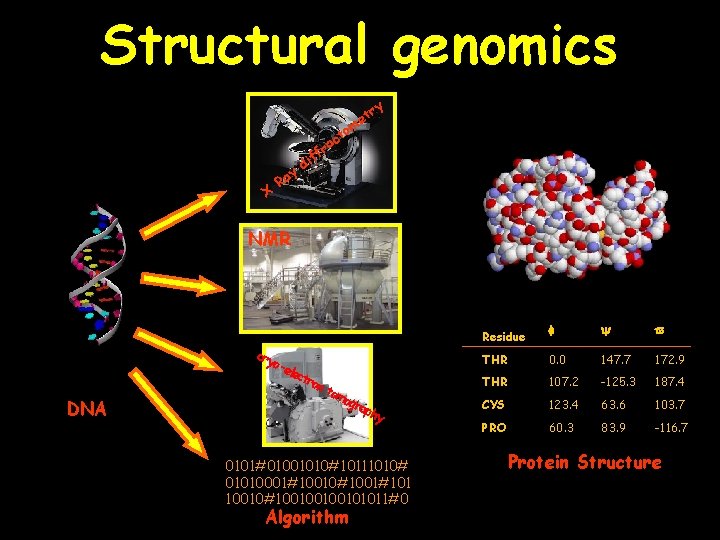
Structural genomics ry t me to X y Ra c ra f if d NMR DNA cry o-e lect ron tom ogr aph y 0101#01001010#10111010# 01010001#10010#1001#101 10010#100100100101011#0 3. genome analysis Algorithm Residue THR 0. 0 147. 7 172. 9 THR 107. 2 -125. 3 187. 4 CYS 123. 4 63. 6 103. 7 PRO 60. 3 83. 9 -116. 7 Protein Structure 16

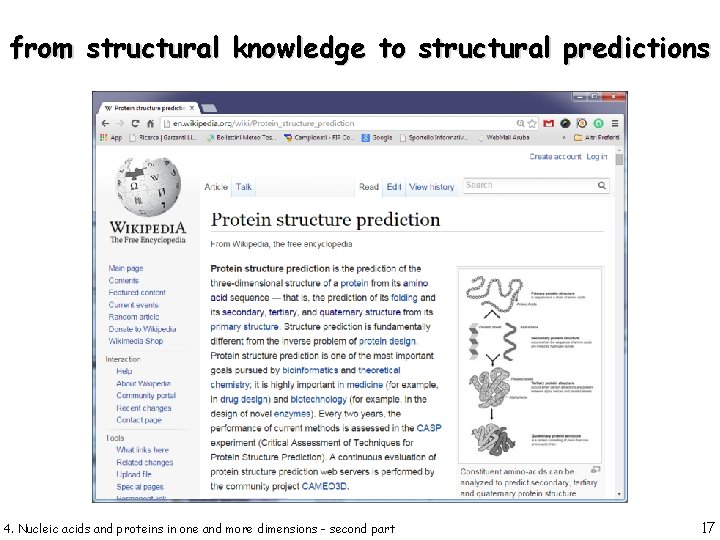
from structural knowledge to structural predictions 4. Nucleic acids and proteins in one and more dimensions - second part 17

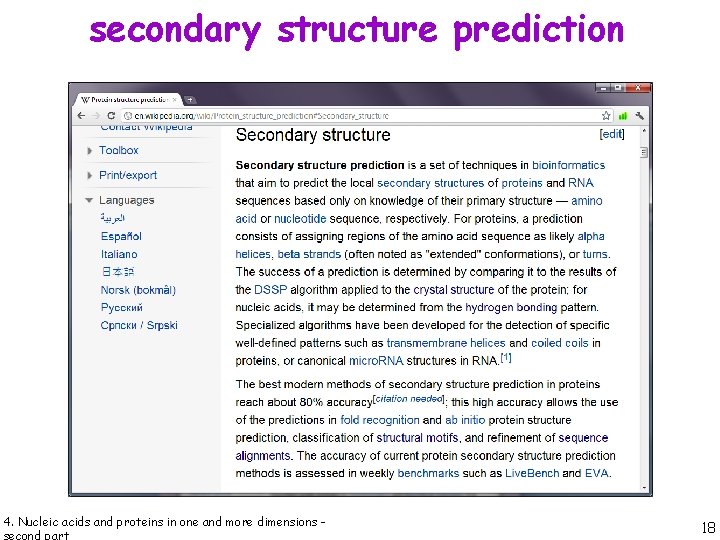
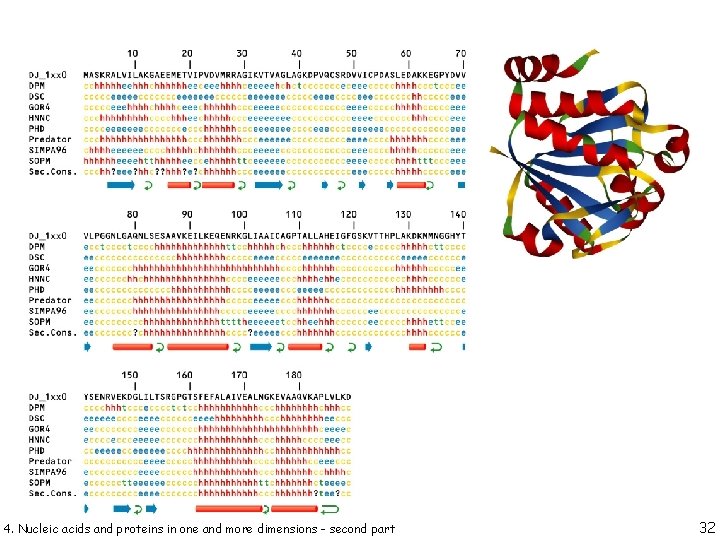
secondary structure prediction 4. Nucleic acids and proteins in one and more dimensions - 18

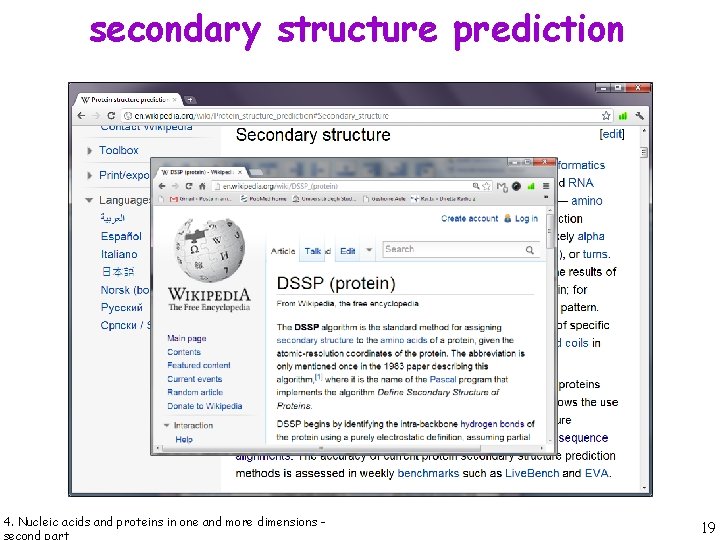
secondary structure prediction 4. Nucleic acids and proteins in one and more dimensions - 19

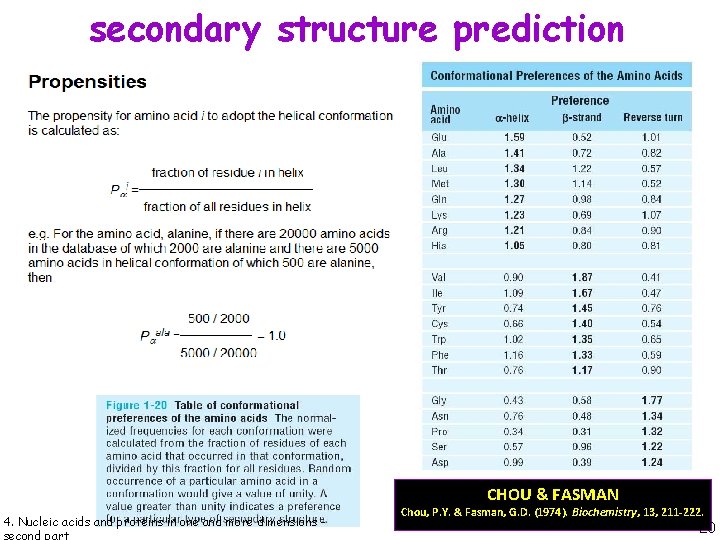
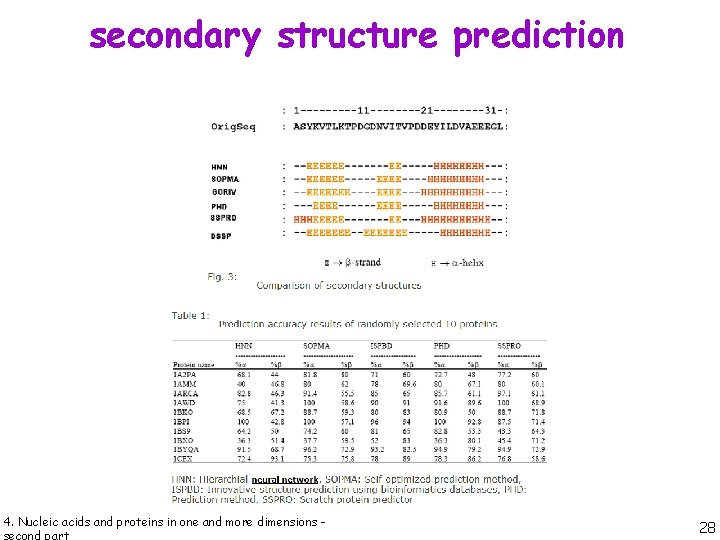
secondary structure prediction CHOU & FASMAN 4. Nucleic acids and proteins in one and more dimensions - Chou, P. Y. & Fasman, G. D. (1974). Biochemistry, 13, 211 -222. 20

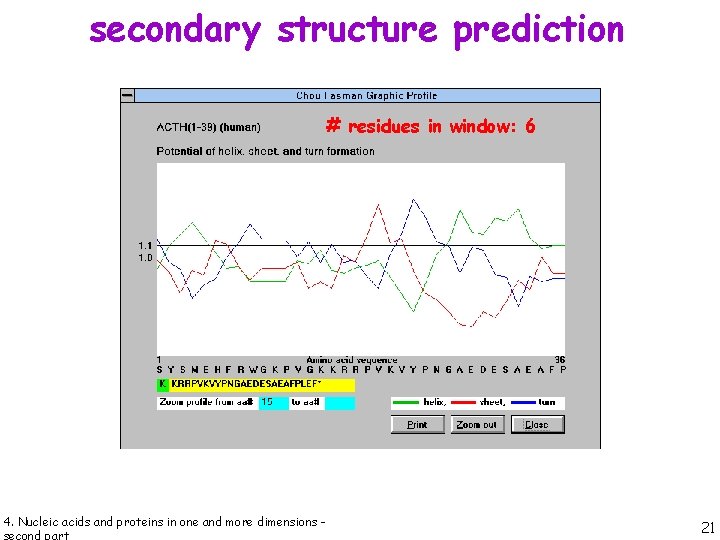
secondary structure prediction # residues in window: 6 4. Nucleic acids and proteins in one and more dimensions - 21

secondary structure prediction 4. Nucleic acids and proteins in one and more dimensions - 22

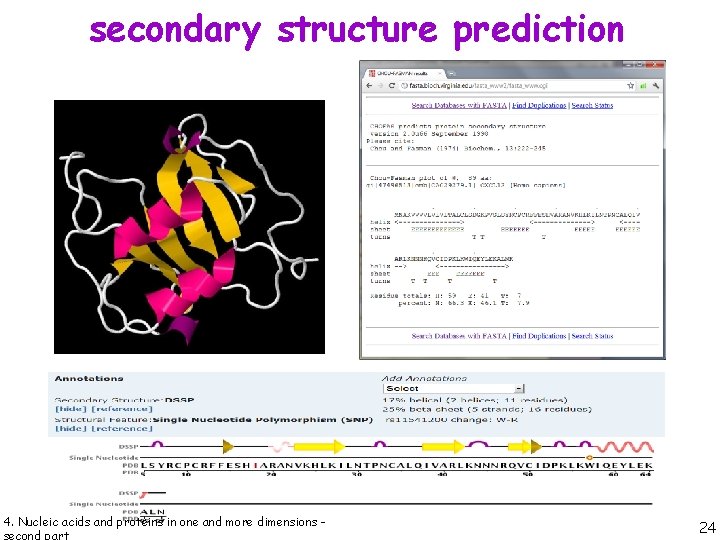
secondary structure prediction 4. Nucleic acids and proteins in one and more dimensions - 23

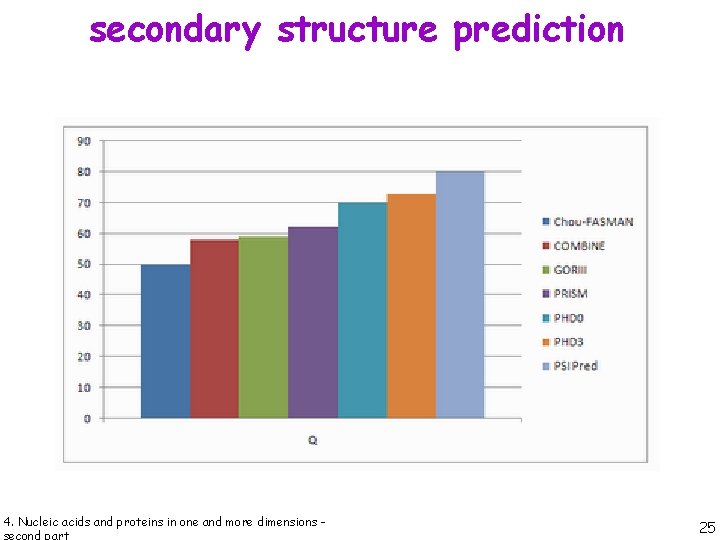
secondary structure prediction 4. Nucleic acids and proteins in one and more dimensions - 24

secondary structure prediction 4. Nucleic acids and proteins in one and more dimensions - 25

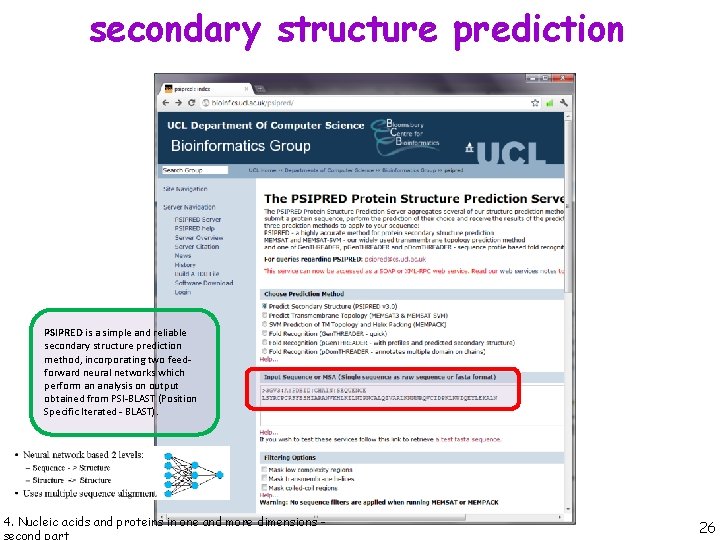
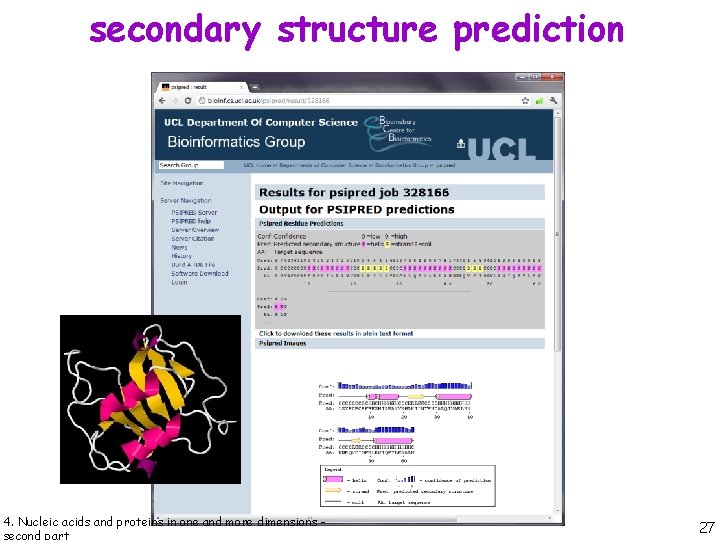
secondary structure prediction PSIPRED is a simple and reliable secondary structure prediction method, incorporating two feedforward neural networks which perform an analysis on output obtained from PSI-BLAST (Position Specific Iterated - BLAST). 4. Nucleic acids and proteins in one and more dimensions - 26

secondary structure prediction 4. Nucleic acids and proteins in one and more dimensions - 27

secondary structure prediction 4. Nucleic acids and proteins in one and more dimensions - 28

4. Nucleic acids and proteins in one and more dimensions - second part 29

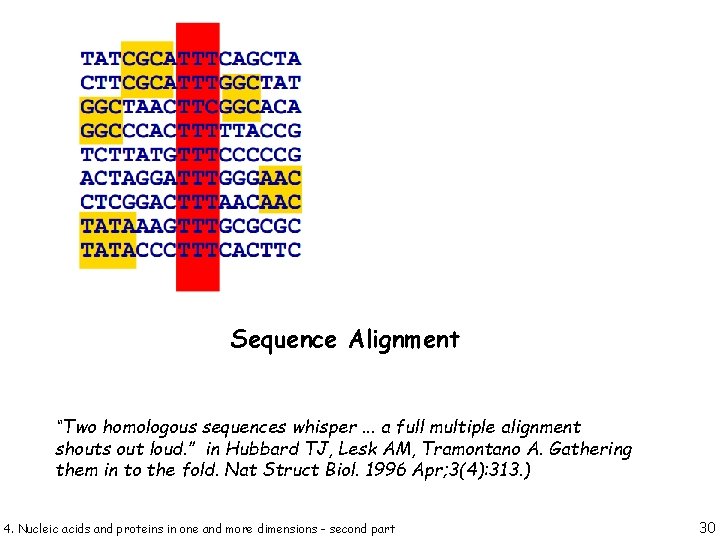
Sequence Alignment “Two homologous sequences whisper. . . a full multiple alignment shouts out loud. ” in Hubbard TJ, Lesk AM, Tramontano A. Gathering them in to the fold. Nat Struct Biol. 1996 Apr; 3(4): 313. ) 4. Nucleic acids and proteins in one and more dimensions - second part 30

4. Nucleic acids and proteins in one and more dimensions - second part 31

4. Nucleic acids and proteins in one and more dimensions - second part 32

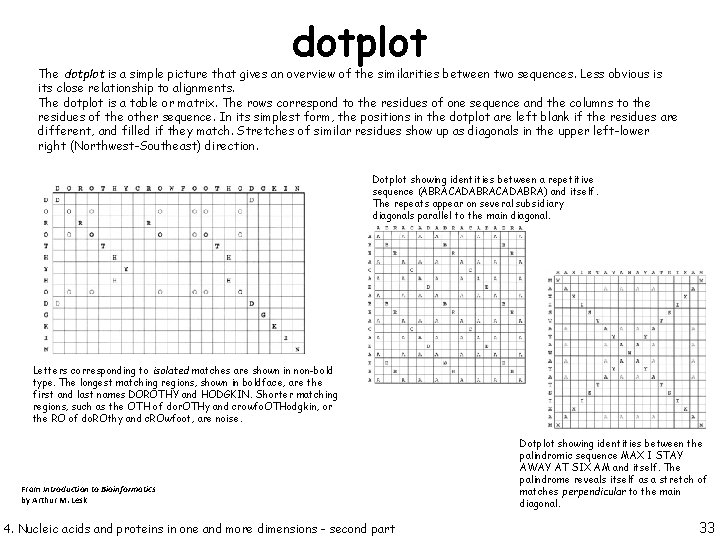
dotplot The dotplot is a simple picture that gives an overview of the similarities between two sequences. Less obvious is its close relationship to alignments. The dotplot is a table or matrix. The rows correspond to the residues of one sequence and the columns to the residues of the other sequence. In its simplest form, the positions in the dotplot are left blank if the residues are different, and filled if they match. Stretches of similar residues show up as diagonals in the upper left-lower right (Northwest-Southeast) direction. Dotplot showing identities between a repetitive sequence (ABRACADABRA) and itself. The repeats appear on several subsidiary diagonals parallel to the main diagonal. Letters corresponding to isolated matches are shown in non-bold type. The longest matching regions, shown in boldface, are the first and last names DOROTHY and HODGKIN. Shorter matching regions, such as the OTH of dor. OTHy and crowfo. OTHodgkin, or the RO of do. ROthy and c. ROwfoot, are noise. From Introduction to Bioinformatics by Arthur M. Lesk 4. Nucleic acids and proteins in one and more dimensions - second part Dotplot showing identities between the palindromic sequence MAX I STAY AWAY AT SIX AM and itself. The palindrome reveals itself as a stretch of matches perpendicular to the main diagonal. 33

http: //vlab. amrita. edu/? sub=3&brch=274&sim=1434&cnt=1 34

4. Nucleic acids and proteins in one and more dimensions - 35

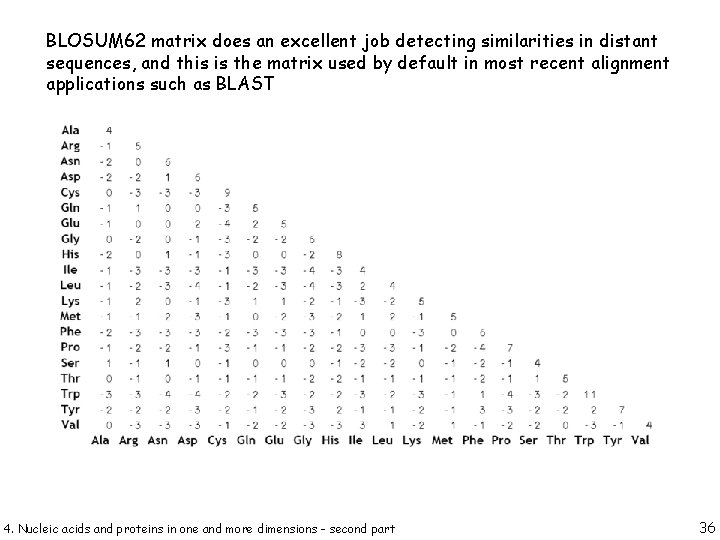
BLOSUM 62 matrix does an excellent job detecting similarities in distant sequences, and this is the matrix used by default in most recent alignment applications such as BLAST 4. Nucleic acids and proteins in one and more dimensions - second part 36

Mutation probability matrix for the evolutionary distance of 250 PAMs 4. Nucleic acids and proteins in one and more dimensions - second part 37

4. Nucleic acids and proteins in one and more dimensions - 38

4. Nucleic acids and proteins in one and more dimensions - 39

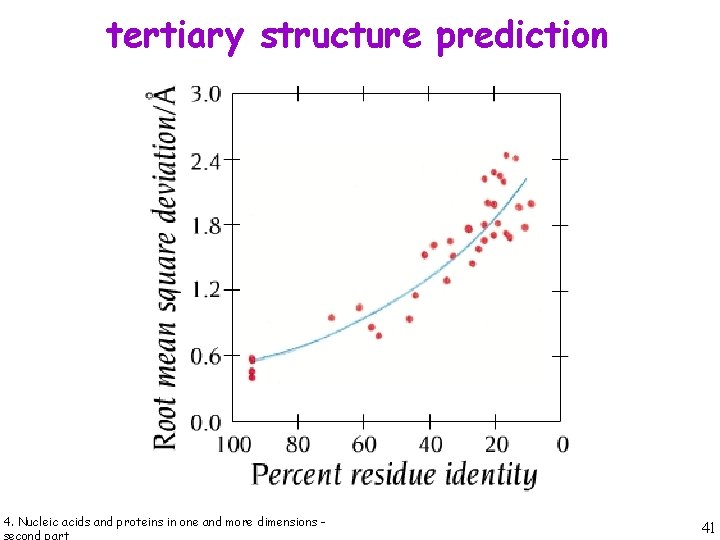
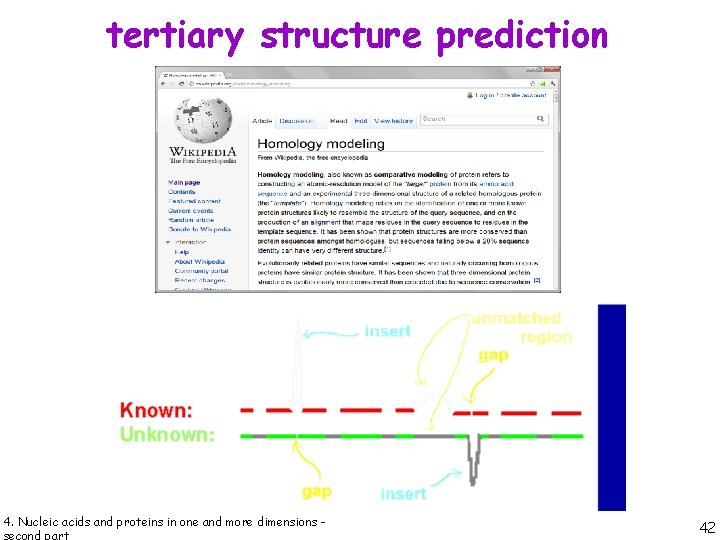
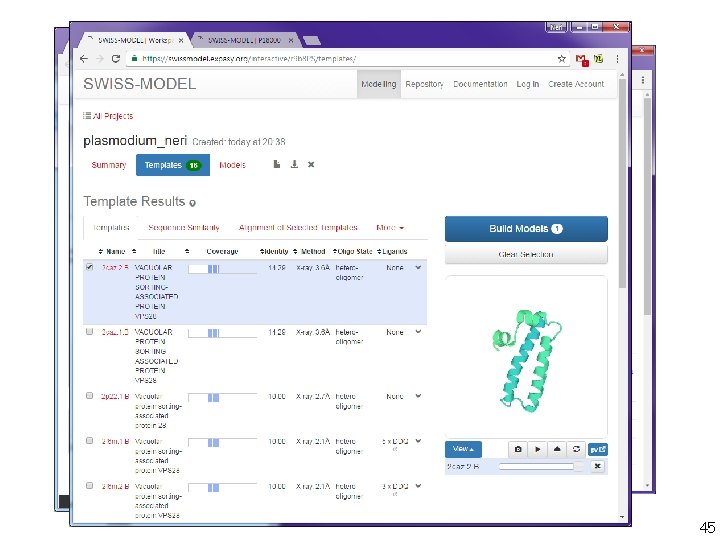
tertiary structure prediction 4. Nucleic acids and proteins in one and more dimensions - 40

tertiary structure prediction 4. Nucleic acids and proteins in one and more dimensions - 41

tertiary structure prediction 4. Nucleic acids and proteins in one and more dimensions - 42

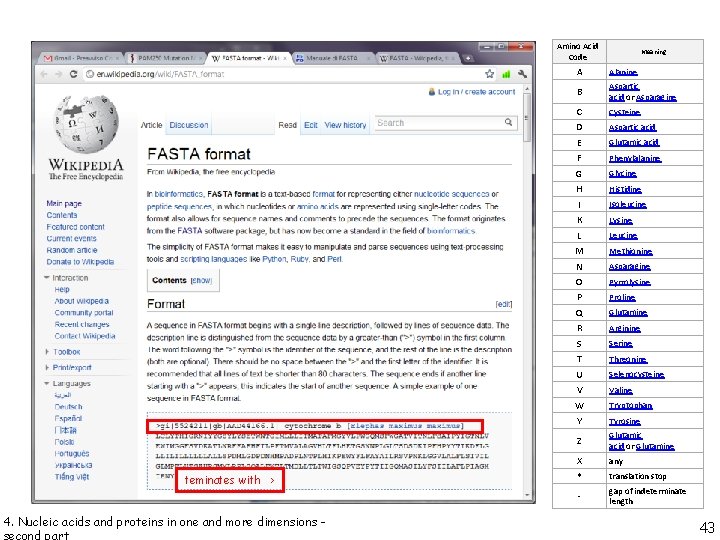
Amino Acid Code teminates with > 4. Nucleic acids and proteins in one and more dimensions - Meaning A Alanine B Aspartic acid or Asparagine C D E F G H I K L M N O P Q R S T U V W Y Cysteine Z Glutamic acid or Glutamine X * any - gap of indeterminate length Aspartic acid Glutamic acid Phenylalanine Glycine Histidine Isoleucine Lysine Leucine Methionine Asparagine Pyrrolysine Proline Glutamine Arginine Serine Threonine Selenocysteine Valine Tryptophan Tyrosine translation stop 43

4. Nucleic acids and proteins in one and more dimensions - 44

45

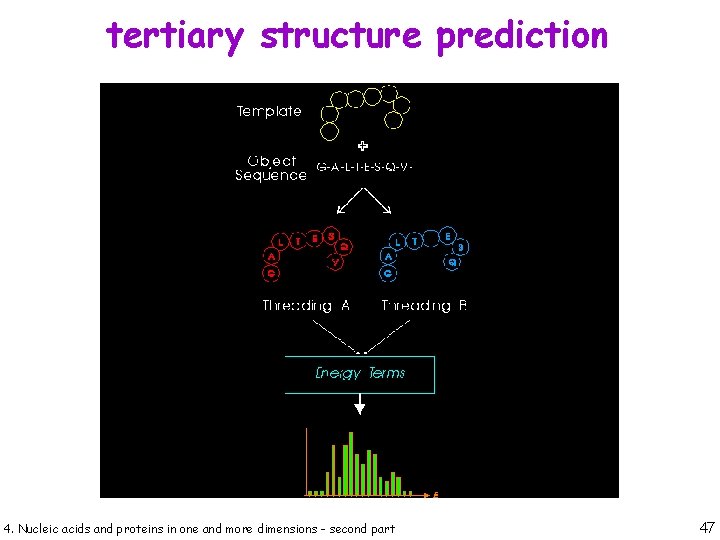
tertiary structure prediction 4. Nucleic acids and proteins in one and more dimensions - second part 46

tertiary structure prediction 4. Nucleic acids and proteins in one and more dimensions - second part 47

tertiary structure prediction 4. Nucleic acids and proteins in one and more dimensions - second part 48

Protein folding ab initio calculations of protein structure 4. Nucleic acids and proteins in one and more dimensions - second part 49

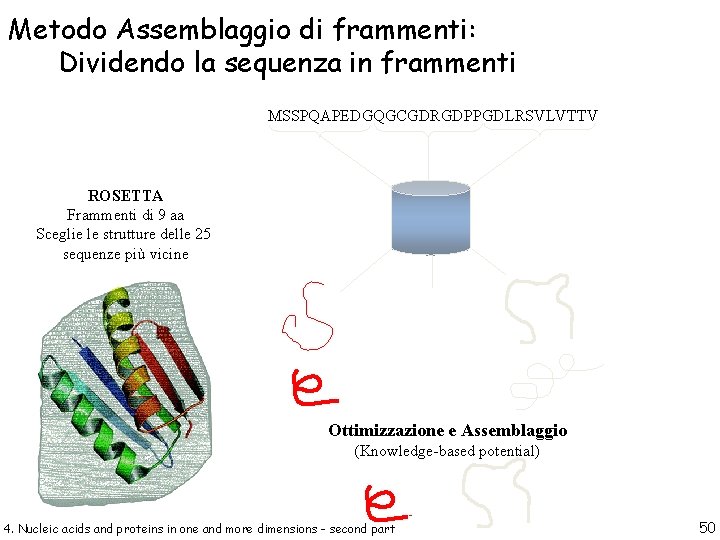
Metodo Assemblaggio di frammenti: Dividendo la sequenza in frammenti MSSPQAPEDGQGCGDRGDPPGDLRSVLVTTV ROSETTA Frammenti di 9 aa Sceglie le strutture delle 25 sequenze più vicine Ottimizzazione e Assemblaggio (Knowledge-based potential) 4. Nucleic acids and proteins in one and more dimensions - second part 50

Rosetta Fragment Libraries n 25 -200 fragments for each 3 and 9 residue sequence window n Selected from database of known structures > 2. 5Å resolution < 50% sequence identity n Ranked by sequence similarity and similarity of predicted and known secondary structure 4. Nucleic acids and proteins in one and more dimensions - second part 51

4. Nucleic acids and proteins in one and more dimensions - second part 52

RNA structure prediction 4. Nucleic acids and proteins in one and more dimensions - second part 53

RNA structure prediction Primary structure of RNA Secondary structure of a telomerase RNA Tertiary structure of RNA 4. Nucleic acids and proteins in one and more dimensions - second part 54

RNA structure prediction 4. Nucleic acids and proteins in one and more dimensions - second part 55