Macromolecules Chapter 5 Macromolecules n Large complex molecules








- Slides: 49

Macromolecules Chapter 5

Macromolecules n Large complex molecules n Carbohydrates, proteins, lipids & nucleic acids

Macromolecules n Polymer n Large molecule n Carboydrates, proteins, nucleic acids n Monomers n Smaller repeating units n Build polymers

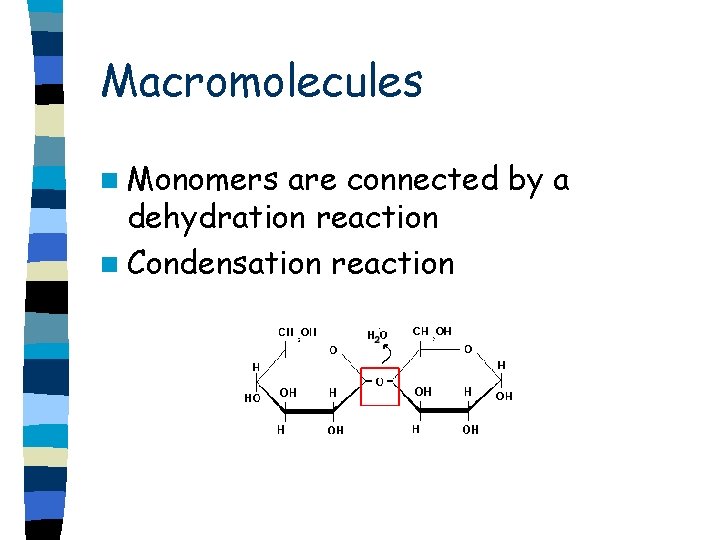
Macromolecules n Monomers are connected by a dehydration reaction n Condensation reaction

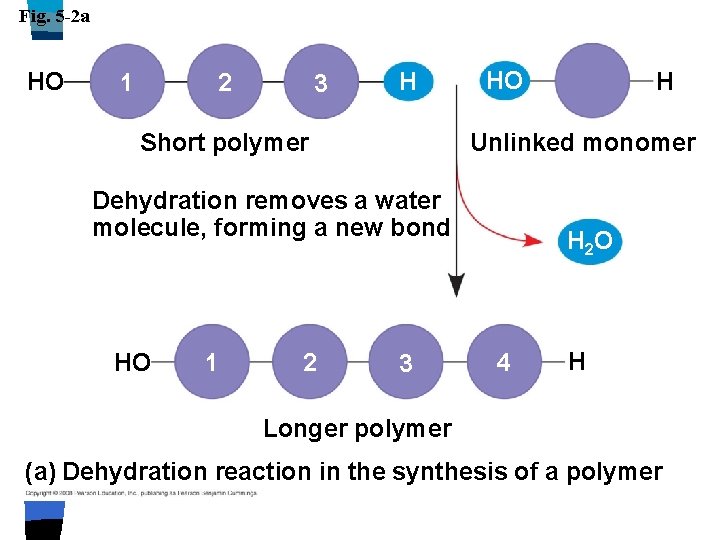
Fig. 5 -2 a HO 1 2 3 H Short polymer HO Unlinked monomer Dehydration removes a water molecule, forming a new bond HO 1 2 H 3 H 2 O 4 H Longer polymer (a) Dehydration reaction in the synthesis of a polymer

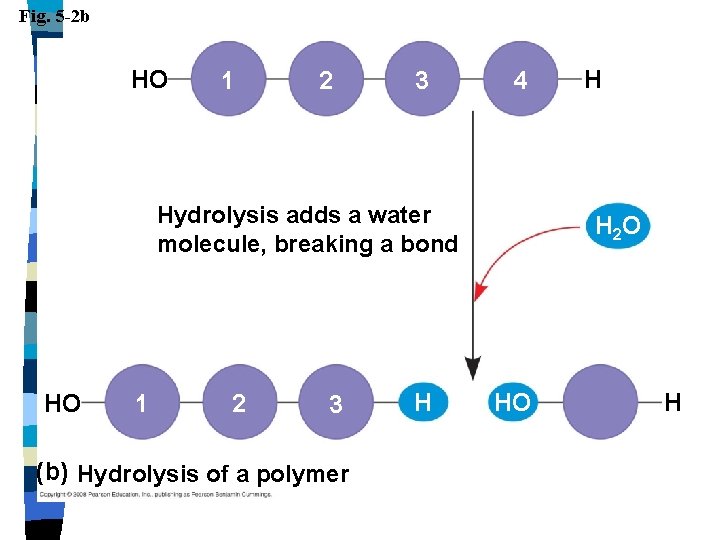
Macromolecules n Polymers are broken apart into monomers by hydrolysis n Bonds are broken by adding water

Fig. 5 -2 b HO 1 2 3 4 Hydrolysis adds a water molecule, breaking a bond HO 1 2 3 (b) Hydrolysis of a polymer H H H 2 O HO H

Carbohydrates n Simple sugars to complex polymers n Stored energy n Structure

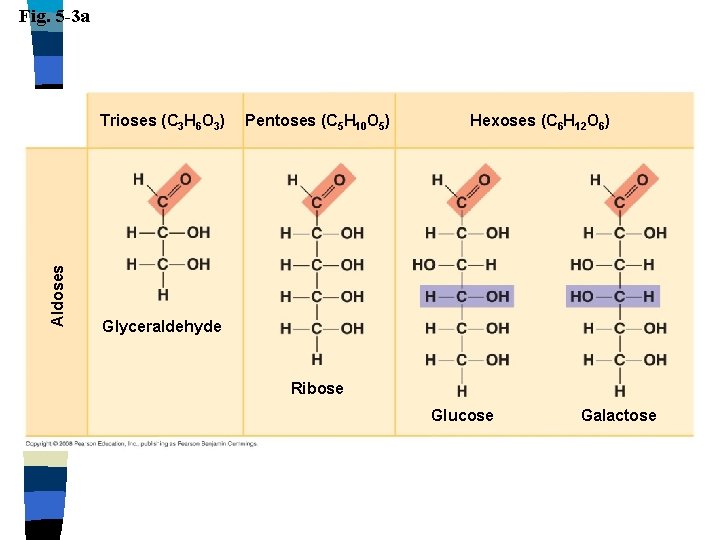
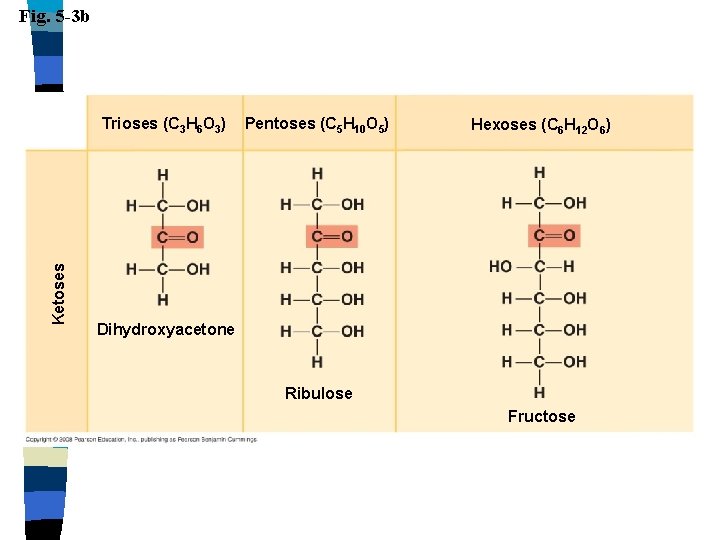
Carbohydrates Monosaccharides n Single sugar n – Glucose, fructose, galactose n n n Molecules contain carbon, hydrogen & oxygen in a 1: 2: 1 ratio CH 2 O End in –ose Aldose: aldehyde sugar Ketose: ketone sugar

Fig. 5 -3 a Aldoses Trioses (C 3 H 6 O 3) Pentoses (C 5 H 10 O 5) Hexoses (C 6 H 12 O 6) Glyceraldehyde Ribose Glucose Galactose

Fig. 5 -3 b Ketoses Trioses (C 3 H 6 O 3) Pentoses (C 5 H 10 O 5) Hexoses (C 6 H 12 O 6) Dihydroxyacetone Ribulose Fructose

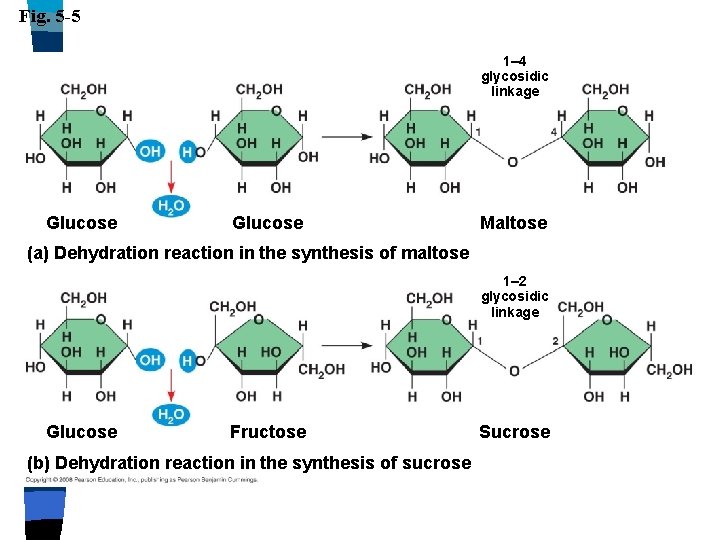
Carbohydrates n Dissaccharides n Two monosaccharides combined n Glycoside linkage: n Covalent bond between two sugars n Sucrose (glucose & fructose) n Maltose (glucose & glucose) n Lactose (glucose & galactose)

Fig. 5 -5 1– 4 glycosidic linkage Glucose Maltose (a) Dehydration reaction in the synthesis of maltose 1– 2 glycosidic linkage Glucose Fructose (b) Dehydration reaction in the synthesis of sucrose Sucrose

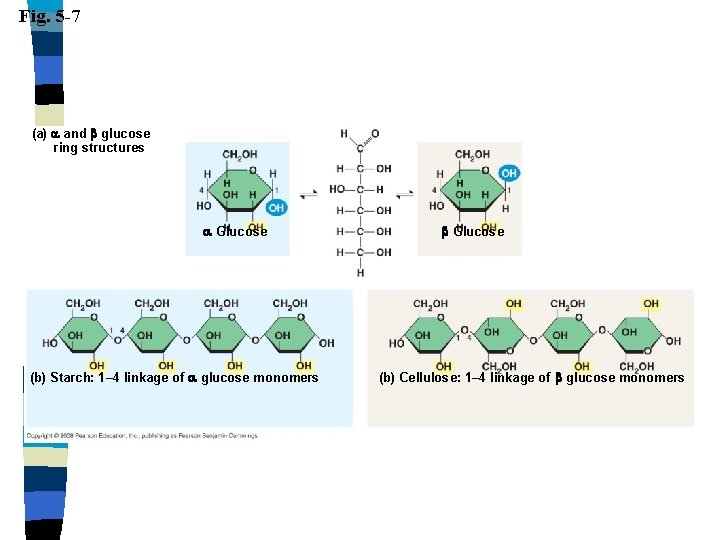
Carbohydrates n Polysaccharides n Many monosaccharides combined n Starch (plant), glycogen (animal), cellulose (plant), chitin

Fig. 5 -7 (a) and glucose ring structures Glucose (b) Starch: 1– 4 linkage of glucose monomers Glucose (b) Cellulose: 1– 4 linkage of glucose monomers


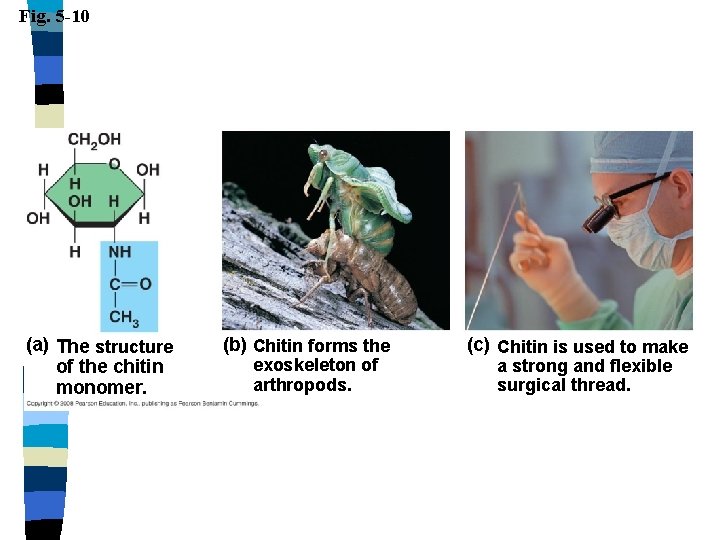
Fig. 5 -10 (a) The structure of the chitin monomer. (b) Chitin forms the exoskeleton of arthropods. (c) Chitin is used to make a strong and flexible surgical thread.

Lipids n Insoluble in water – Hydrophobic n Store energy n Make membranes

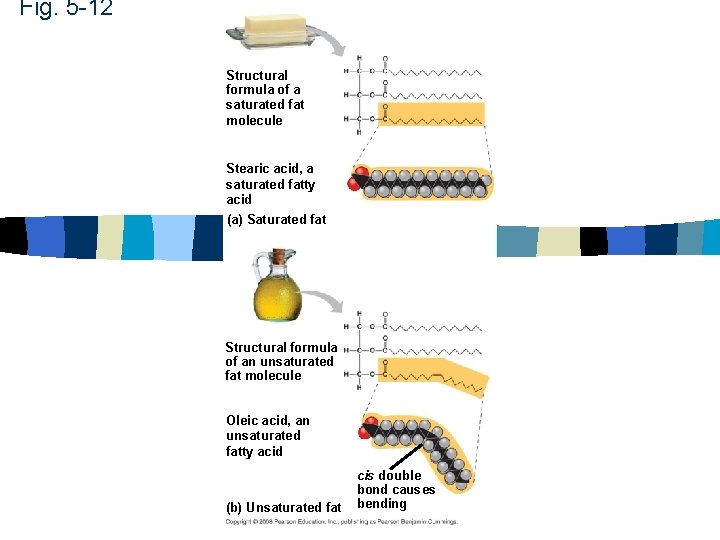
Lipids n Fats – Unsaturated – Saturated n Phospholipids – Glycerol, phosphate, fatty acid n Steroids

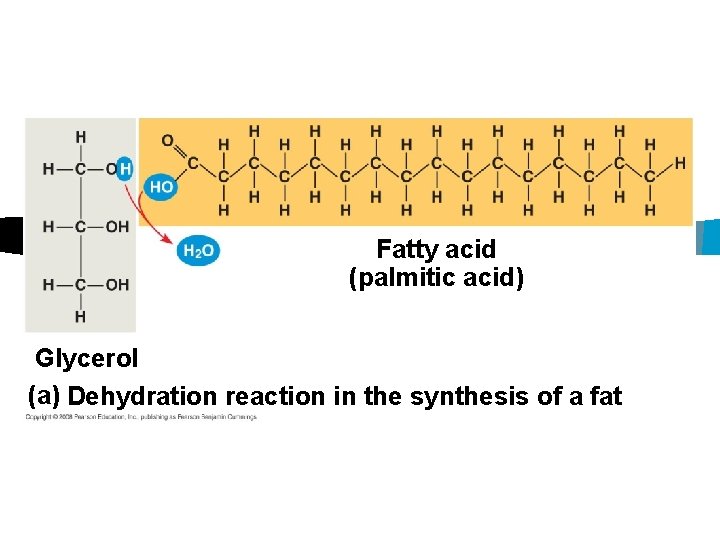
Fats n Glycerol and fatty acids n Triglyceride n 3 fatty acids attached to a glycerol

Fatty acid (palmitic acid) Glycerol (a) Dehydration reaction in the synthesis of a fat

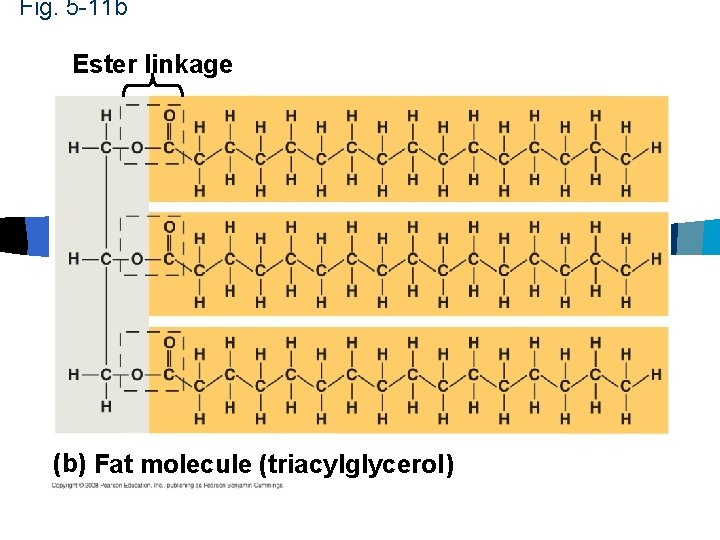
Fig. 5 -11 b Ester linkage (b) Fat molecule (triacylglycerol)

Fig. 5 -12 Structural formula of a saturated fat molecule Stearic acid, a saturated fatty acid (a) Saturated fat Structural formula of an unsaturated fat molecule Oleic acid, an unsaturated fatty acid (b) Unsaturated fat cis double bond causes bending

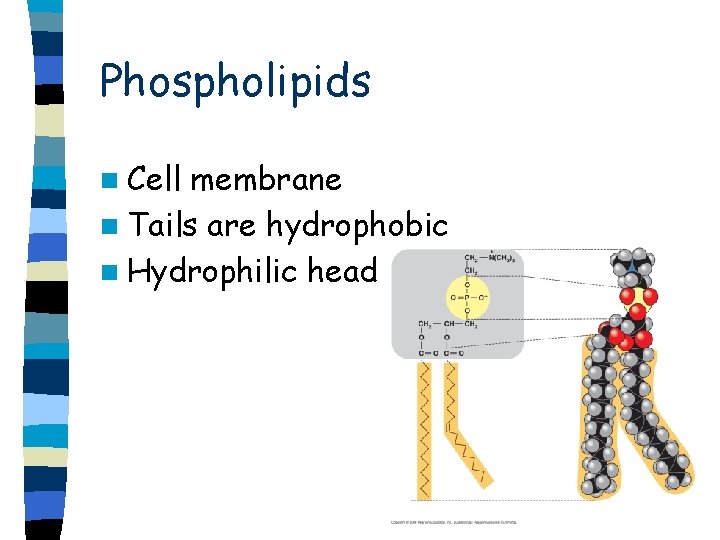
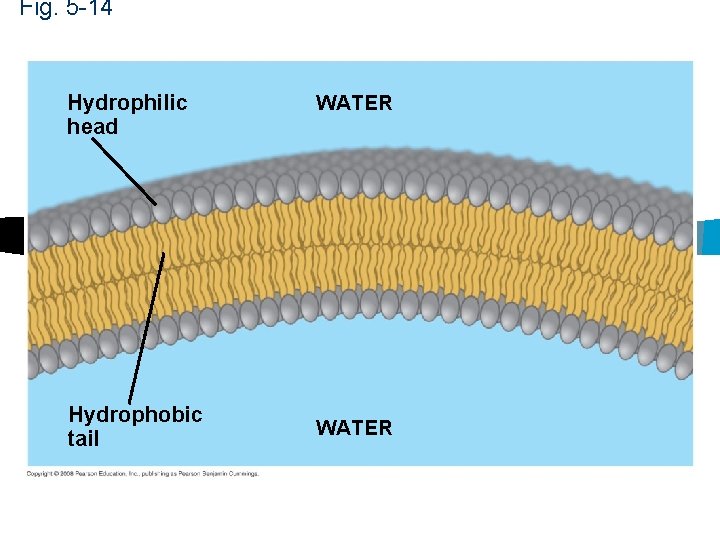
Phospholipids n Cell membrane n Tails are hydrophobic n Hydrophilic head

Fig. 5 -14 Hydrophilic head Hydrophobic tail WATER

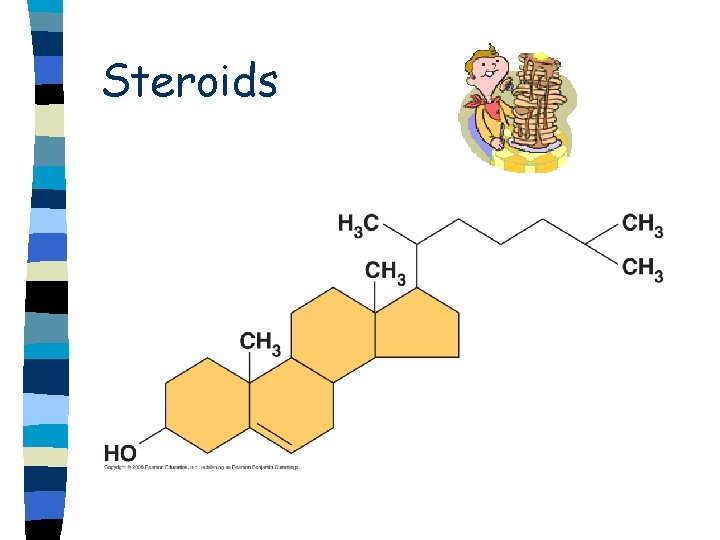
Steroids

Steroids

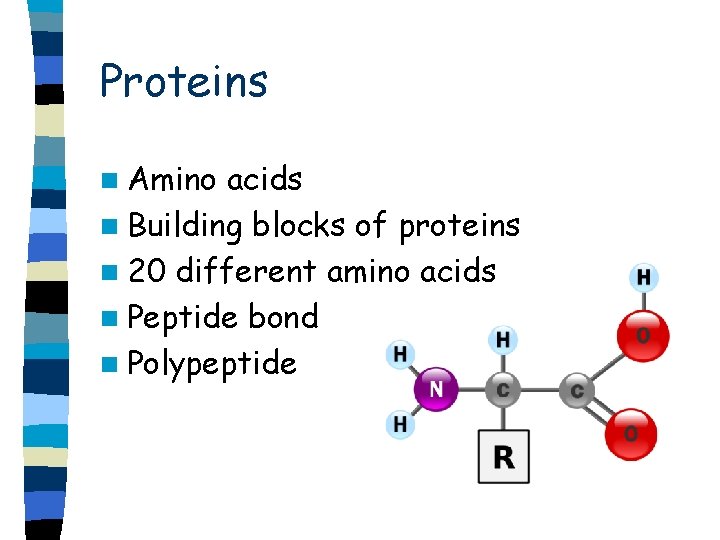
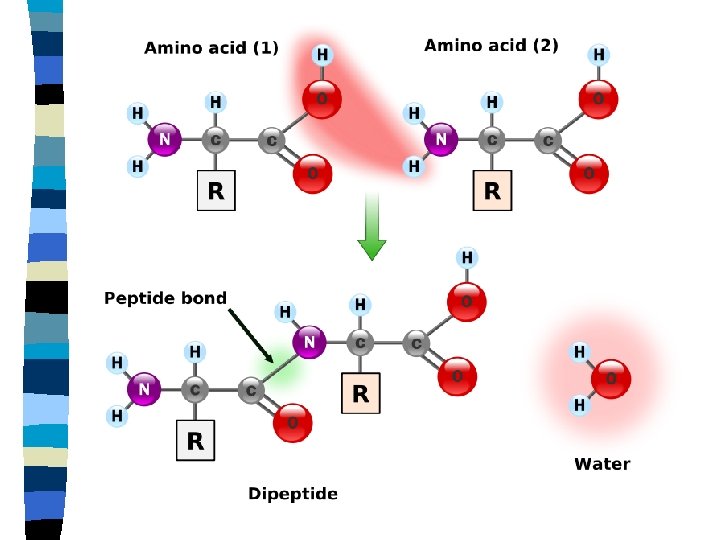
Proteins n Amino acids n Building blocks of proteins n 20 different amino acids n Peptide bond n Polypeptide


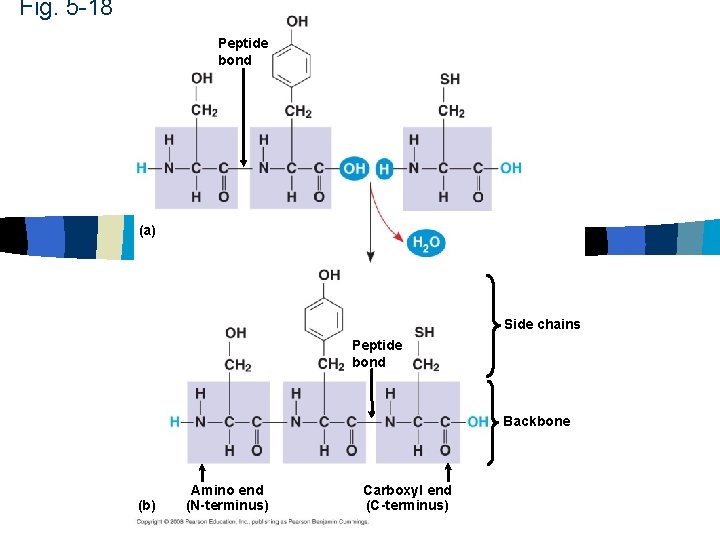
Fig. 5 -18 Peptide bond (a) Side chains Peptide bond Backbone (b) Amino end (N-terminus) Carboxyl end (C-terminus)

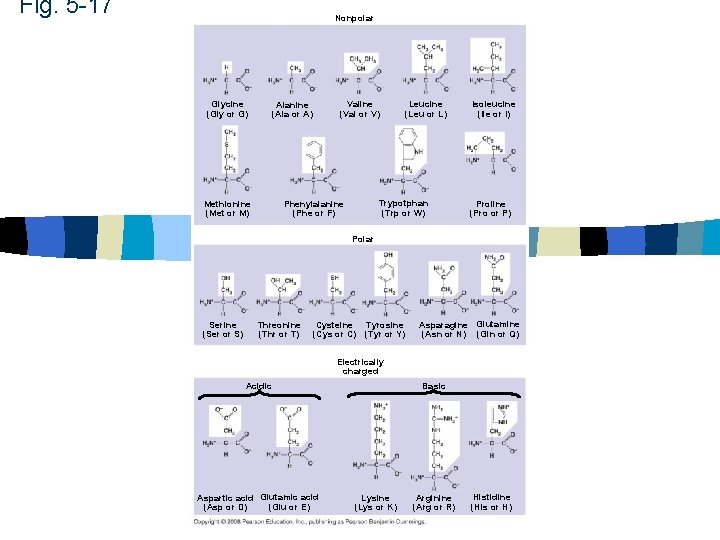
Fig. 5 -17 Nonpolar Glycine (Gly or G) Valine (Val or V) Alanine (Ala or A) Methionine (Met or M) Leucine (Leu or L) Trypotphan (Trp or W) Phenylalanine (Phe or F) Isoleucine (Ile or I) Proline (Pro or P) Polar Serine (Ser or S) Threonine (Thr or T) Cysteine (Cys or C) Tyrosine (Tyr or Y) Asparagine Glutamine (Asn or N) (Gln or Q) Electrically charged Acidic Aspartic acid Glutamic acid (Glu or E) (Asp or D) Basic Lysine (Lys or K) Arginine (Arg or R) Histidine (His or H)

Protein Function n Enzyme catalysis n Defense n Transport n Support n Motion n Regulation n Storage

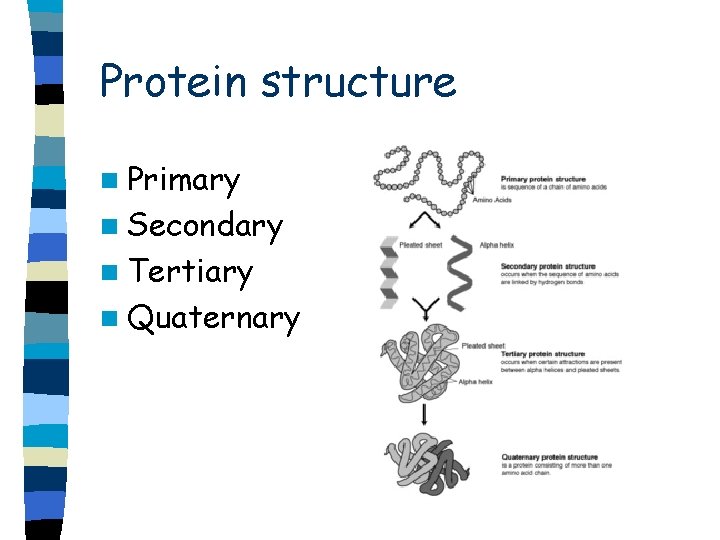
Protein structure n Primary n Secondary n Tertiary n Quaternary

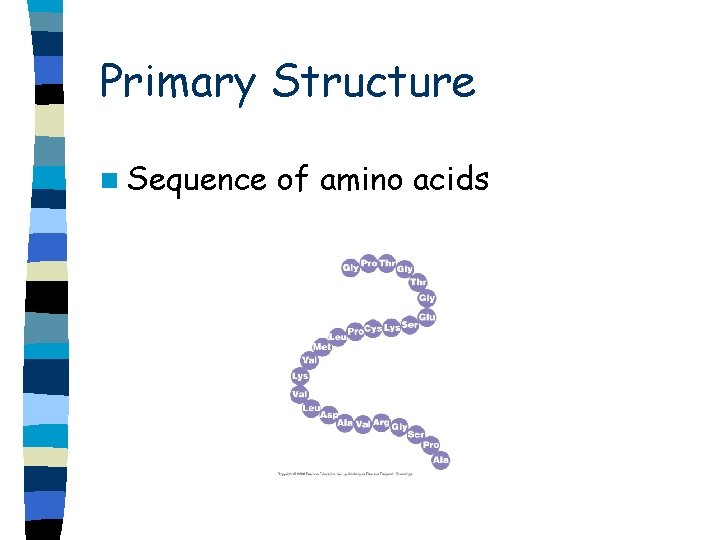
Primary Structure n Sequence of amino acids

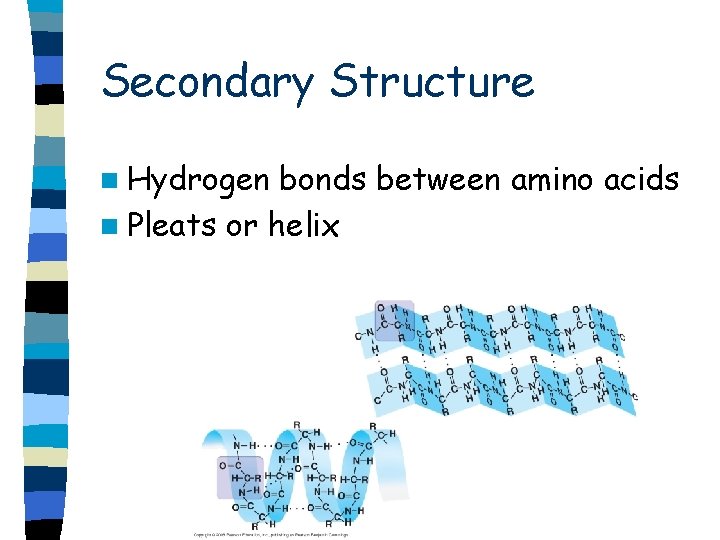
Secondary Structure n Hydrogen bonds between amino acids n Pleats or helix

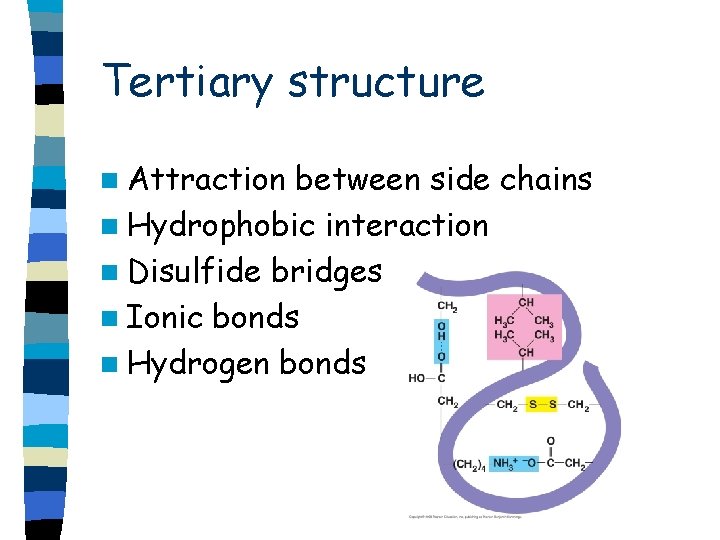
Tertiary structure n Attraction between side chains n Hydrophobic interaction n Disulfide bridges n Ionic bonds n Hydrogen bonds

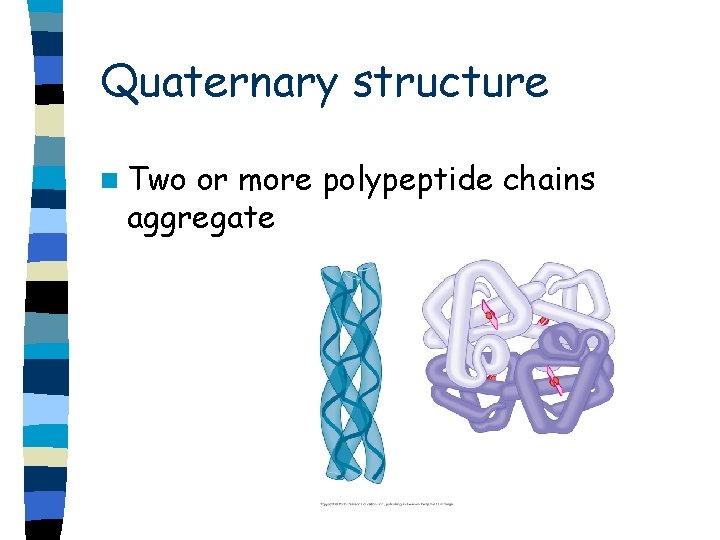
Quaternary structure n Two or more polypeptide chains aggregate

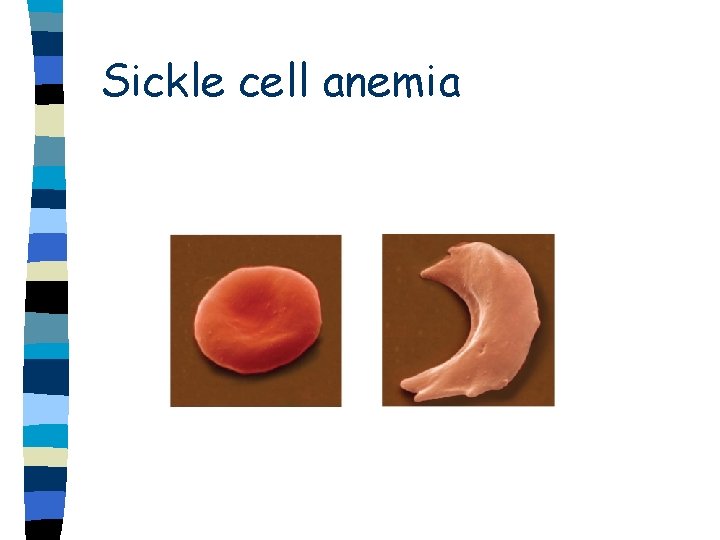
Sickle cell anemia

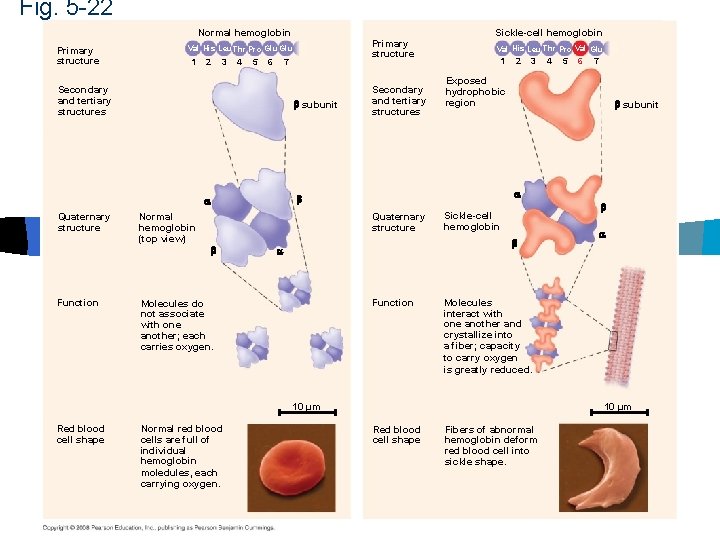
Fig. 5 -22 Normal hemoglobin Primary structure 1 2 3 4 5 6 7 Secondary and tertiary structures subunit Function Normal hemoglobin (top view) Secondary and tertiary structures Val His Leu Thr Pro Val Glu 1 2 3 Normal red blood cells are full of individual hemoglobin moledules, each carrying oxygen. 6 7 subunit Sickle-cell hemoglobin Function Molecules interact with one another and crystallize into a fiber; capacity to carry oxygen is greatly reduced. 10 µm Red blood cell shape 5 Exposed hydrophobic region Molecules do not associate with one another; each carries oxygen. 4 Quaternary structure Sickle-cell hemoglobin Quaternary structure Primary structure Val His Leu Thr Pro Glu 10 µm Red blood cell shape Fibers of abnormal hemoglobin deform red blood cell into sickle shape.

Structure n Denaturation: n Alter, unravel shape of protein n Temperature, p. H, salt n Chaperonins: n Proteins that help with structure

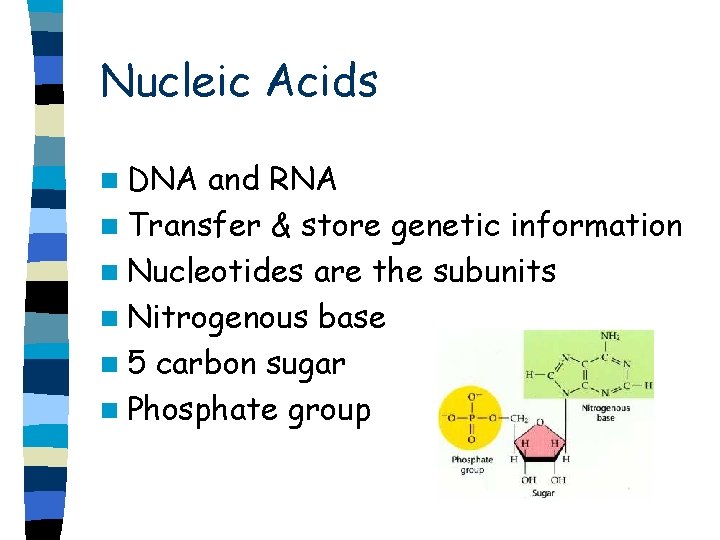
Nucleic Acids n DNA and RNA n Transfer & store genetic information n Nucleotides are the subunits n Nitrogenous base n 5 carbon sugar n Phosphate group

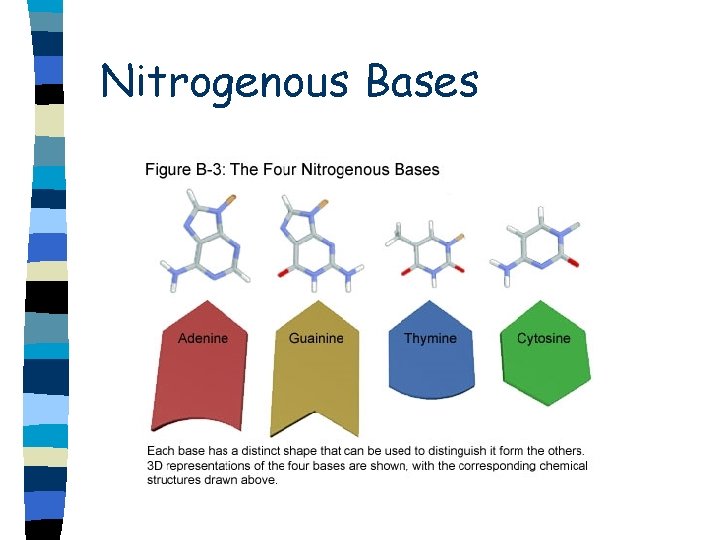
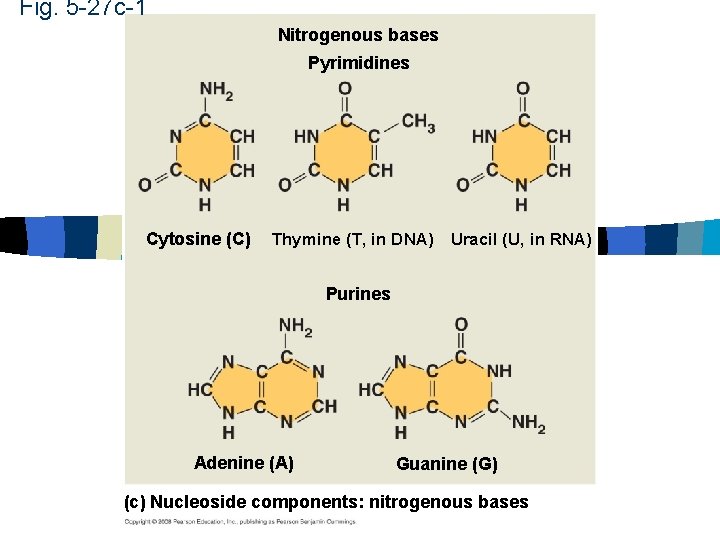
Nucleic Acids n Pyrimidines n Cytosine, thymine and uracil n Single carbon ring n Purines n Adenine, guanine n Double ring structure

Nitrogenous Bases

Fig. 5 -27 c-1 Nitrogenous bases Pyrimidines Cytosine (C) Thymine (T, in DNA) Uracil (U, in RNA) Purines Adenine (A) Guanine (G) (c) Nucleoside components: nitrogenous bases

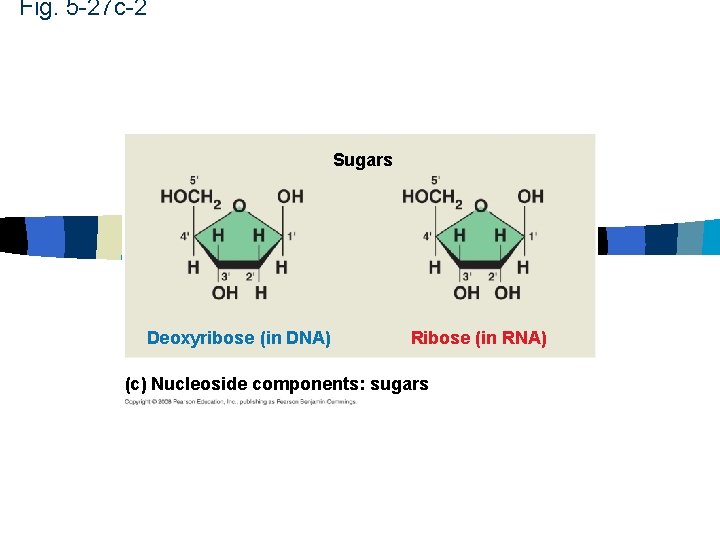
Fig. 5 -27 c-2 Sugars Deoxyribose (in DNA) Ribose (in RNA) (c) Nucleoside components: sugars

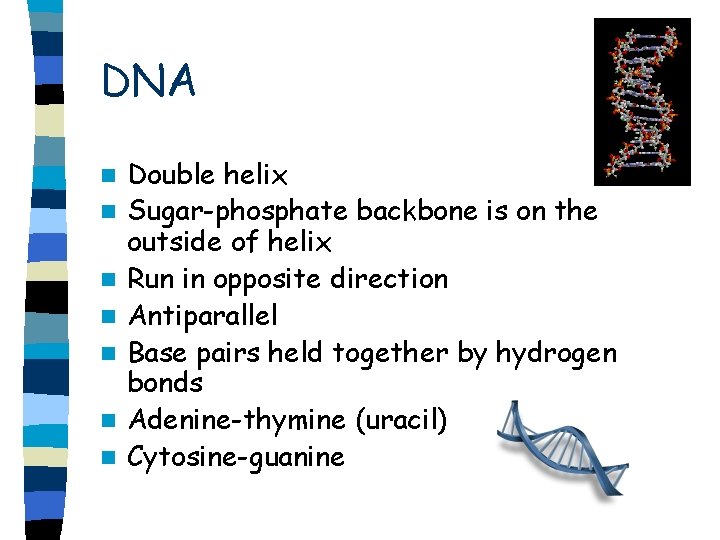
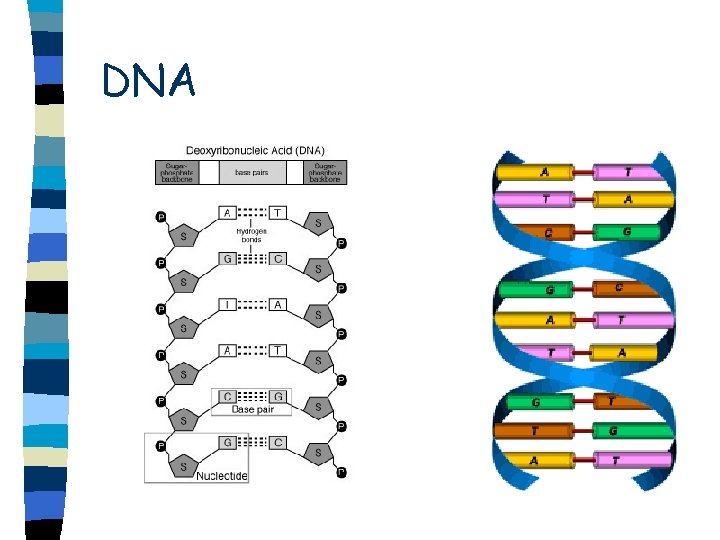
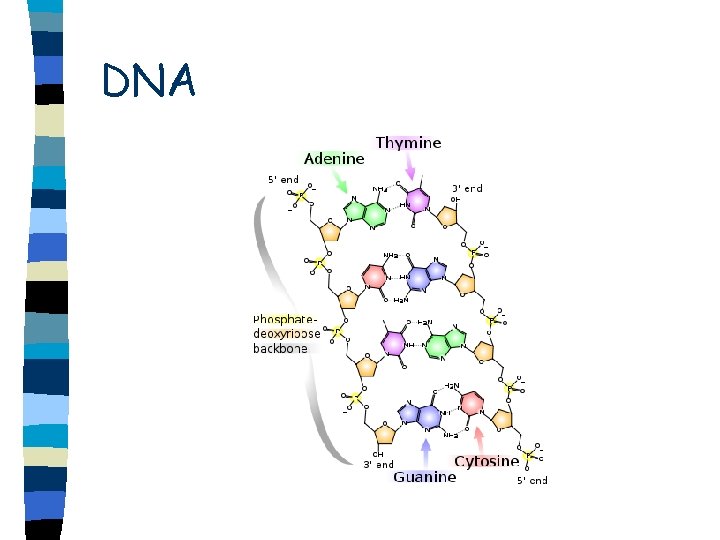
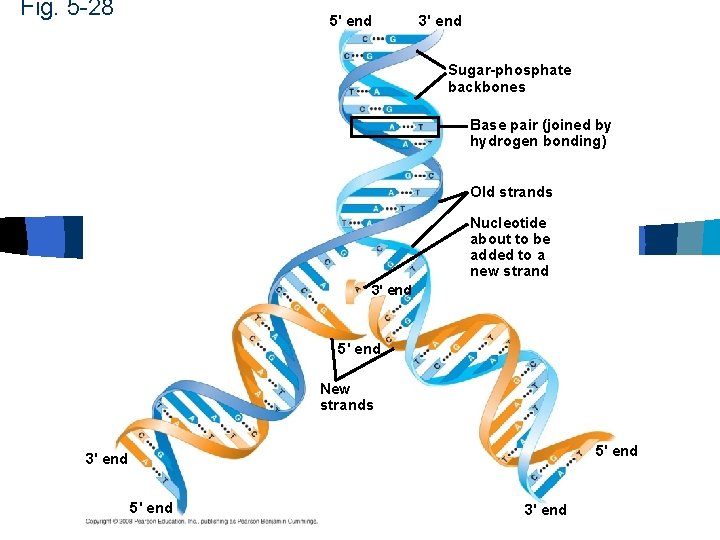
DNA n n n n Double helix Sugar-phosphate backbone is on the outside of helix Run in opposite direction Antiparallel Base pairs held together by hydrogen bonds Adenine-thymine (uracil) Cytosine-guanine

DNA

DNA

Fig. 5 -28 5' end 3' end Sugar-phosphate backbones Base pair (joined by hydrogen bonding) Old strands Nucleotide about to be added to a new strand 3' end 5' end New strands 5' end 3' end