Macromolecular Structure Molecules Andy Howard Biology 555 Fall
Macromolecular Structure: Molecules Andy Howard Biology 555, Fall 2018 23 August 2018

Prominent secondary structures helix 08/23/2018 antiparallel sheet polyproline helix Macromolecular Structure: Molecules p. 2 of 42
- helix • • • Right handed helix, 3. 6 residues/turn 1. 5 Å rise/subunit (18/5 helix) Net dipole moment from ends, others cancel H-bond from NH to carbonyl 4 residues earlier Steric considerations favor formation of H-bonds between amide proton and carbonyl oxygen Other helical forms exist, including: • 310 helix (H-bonds 3 residues earlier) • helix (H-bonds 5 residues earlier) 08/23/2018 Macromolecular Structure: Molecules p. 3 of 42

Contrasting and 310 helices • 08/23/2018 310 helix has 3 residues per turn and displays 31 symmetry Macromolecular Structure: Molecules p. 4 of 42
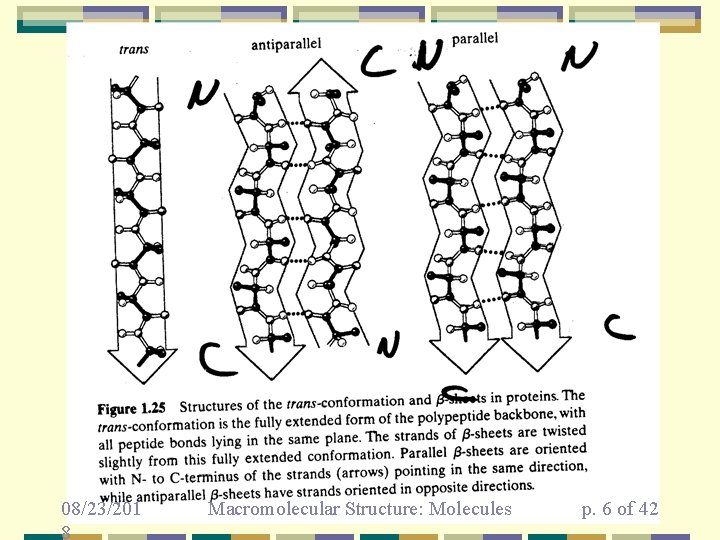
-sheets • • • more extended structure parallel and anti-parallel forms stabilized H-bonds between strands isolated strands are rare strands can be quite far apart in sequence 08/23/2018 Macromolecular Structure: Molecules p. 5 of 42
08/23/201 8 Macromolecular Structure: Molecules p. 6 of 42
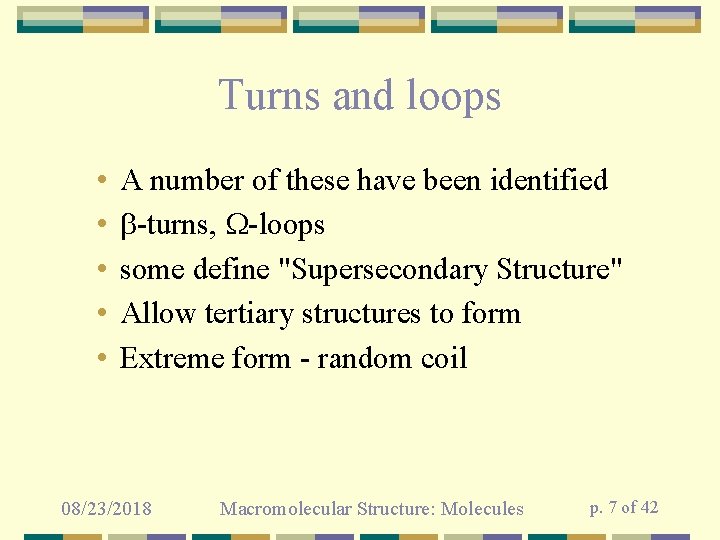
Turns and loops • • • A number of these have been identified -turns, -loops some define "Supersecondary Structure" Allow tertiary structures to form Extreme form - random coil 08/23/2018 Macromolecular Structure: Molecules p. 7 of 42
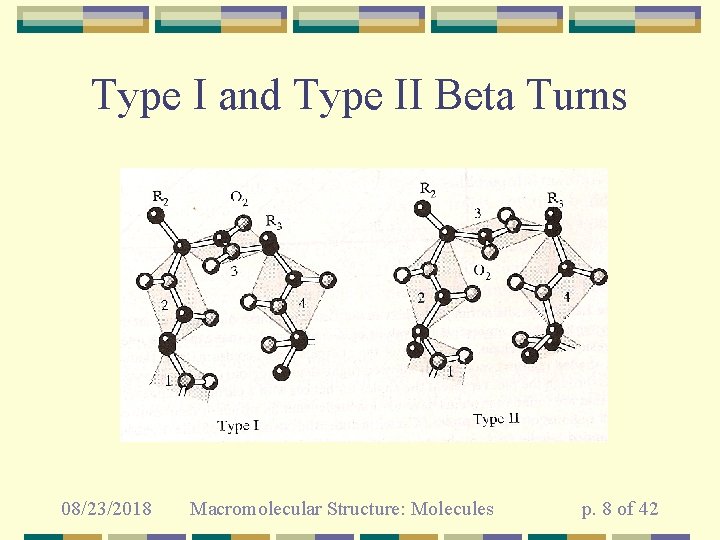
Type I and Type II Beta Turns 08/23/2018 Macromolecular Structure: Molecules p. 8 of 42
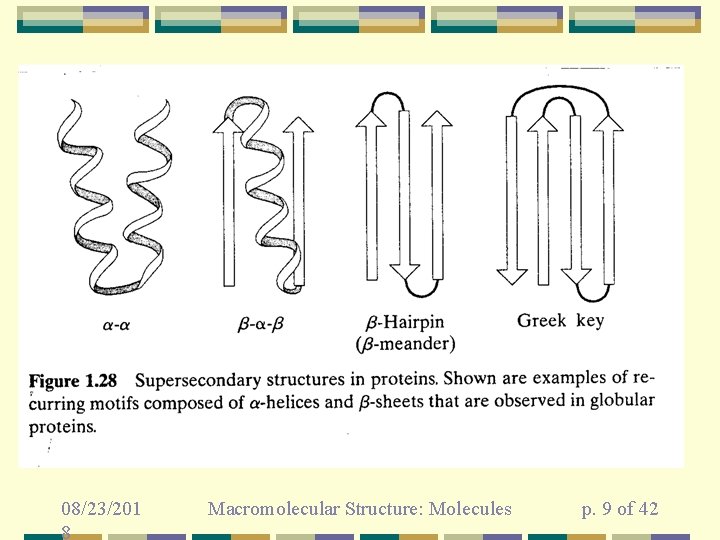
08/23/201 8 Macromolecular Structure: Molecules p. 9 of 42
Tertiary Structure • • • Stabilized mainly by weak interactions Generally have hydrophobic interactions between the side chains on secondary structures Secondary and tertiary structures likely highly interdependent and folding (in the dynamic sense!) is a highly cooperative process 08/23/2018 Macromolecular Structure: Molecules p. 10 of 42
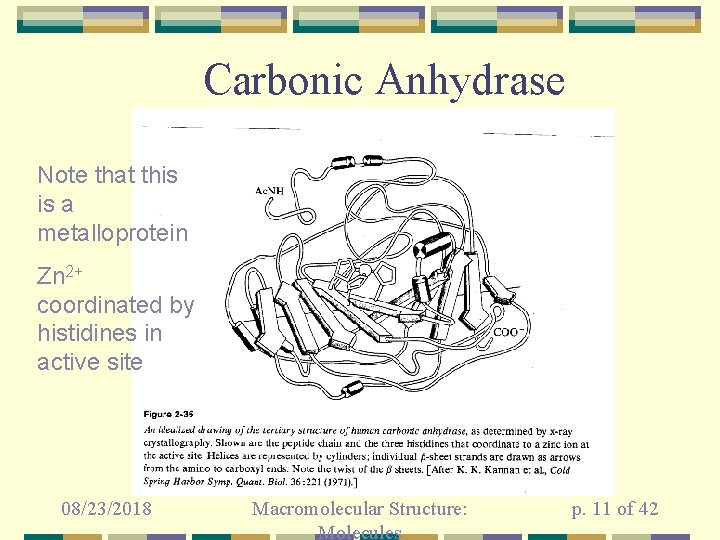
Carbonic Anhydrase Note that this is a metalloprotein Zn 2+ coordinated by histidines in active site 08/23/2018 Macromolecular Structure: Molecules p. 11 of 42
Domains • • Usually defined in functional terms "calcium binding domains", “NAD -binding motifs”, etc. Sometimes identified as fragments made by limited proteolysis Flexible regions assumed to have functional significance. 08/23/2018 Macromolecular Structure: Molecules p. 12 of 42

Visualizing tertiary structure • • • Crystallography generates a list of atoms and coordinates NMR generates a family of possible conformations We then need to be able to examine them: • stick models • CPK models • Ribbon models • All are useful depending on what you are looking for 08/23/2018 Macromolecular Structure: Molecules p. 13 of 42

23/2018 r e Macromolecular Structure: Molecules p. 14
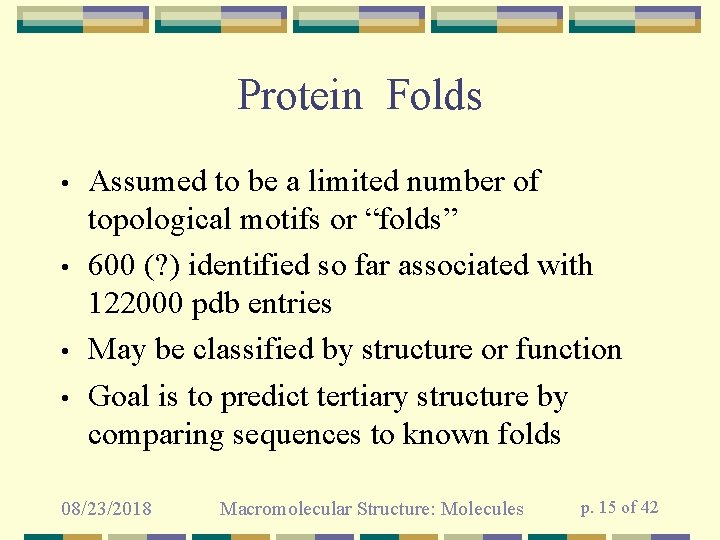
Protein Folds • • Assumed to be a limited number of topological motifs or “folds” 600 (? ) identified so far associated with 122000 pdb entries May be classified by structure or function Goal is to predict tertiary structure by comparing sequences to known folds 08/23/2018 Macromolecular Structure: Molecules p. 15 of 42

Rossmann fold • Pair of motifs with middle shared 08/23/2018 Macromolecular Structure: Molecules p. 16 of 42
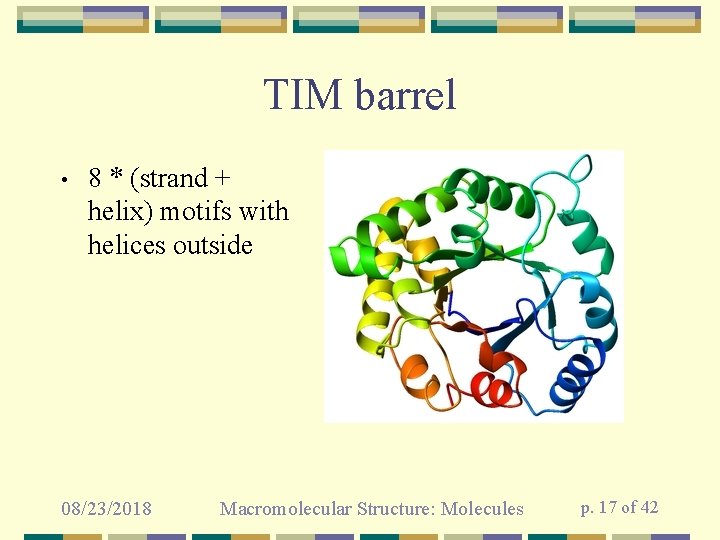
TIM barrel • 8 * (strand + helix) motifs with helices outside 08/23/2018 Macromolecular Structure: Molecules p. 17 of 42

How much can we learn about function from the overall fold? • • • Depends on which overall fold we see Rossmann fold almost always involves binding to NAD, so that probably means a oxidoreductase enzyme TIM barrels get used for all sorts of enzymatic functions, but (as far as I know) they’re always in enzymes 08/23/2018 Macromolecular Structure: Molecules p. 18 of 42
Quaternary structure • • Association of subunits homodimers & heterodimers, tetramers etc. Tend to form regular and symmetric clusters (homodimers, tetramers, …) Maximizes number of energetically favorable interactions with minimum number of sites 08/23/2018 Macromolecular Structure: Molecules p. 19 of 42
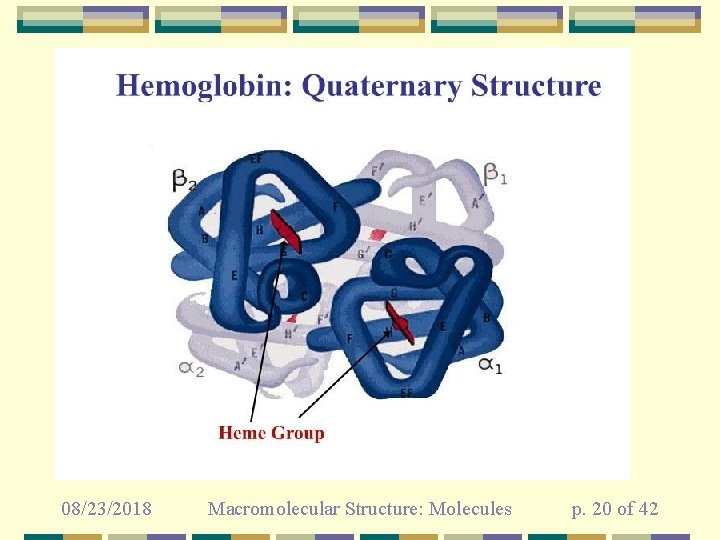
08/23/2018 Macromolecular Structure: Molecules p. 20 of 42
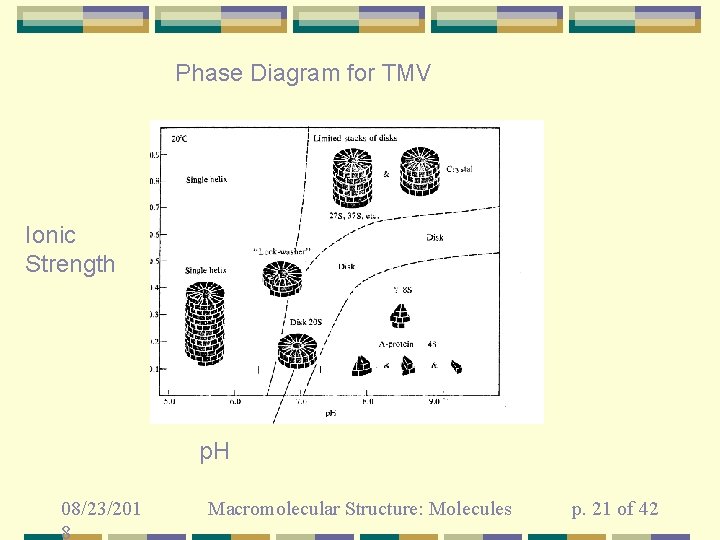
Phase Diagram for TMV Ionic Strength p. H 08/23/201 8 Macromolecular Structure: Molecules p. 21 of 42
Molecular Interactions • Configuration due to covalent bonding • Conformation due to various non-covalent i. e. weak interactions • Interactions may be between components of the macromolecule itself • or between the macromolecule and the surrounding environment • Both very important in determining final 3 -D structure 08/23/2018 Macromolecular Structure: Molecules p. 22 of 42

Weak Interactions • Act to minimize the free energy of a molecule • Differ in the way they depend on distance • Long range • Short range • Energy Dependence: • Charge-charge - 1/r • Charge -dipole - 1/r 2 • Dipole-dipole - 1/r 3 • Charge - induced dipole - 1/r 4 • Induced dipole - induced dipole - 1/r 6 08/23/2018 Macromolecular Structure: Molecules p. 23 of 42
Electrostatic interactions • • • Coulomb’s law: F = q 1 q 2 r -2 / (4 0) … this version is correct if there is a vacuum between the charges q 1 and q 2. If there’s a dielectric in between, then it’s F = q 1 q 2 r -2 / (4 D), Where D is a measure of the amount of charge shielding provided by the medium Water has D = 80 0 Note that energy ( = force distance) is inversely proportional to r, I. e. E ~ r-1 08/23/2018 Macromolecular Structure: Molecules p. 24 of 42
Van der Waals Radius • • • Steric repulsion (r-12 dependence) and Dispersion forces (r-6 dependence) define an interaction energy minimum and a distance of closest approach for two neutral atoms Half this distance called the van der Waals radius Unique for each type of atom If atoms are closer than the sum of the vd. W radii they can be said to be sterically hindered 08/23/2018 Macromolecular Structure: Molecules p. 25 of 42
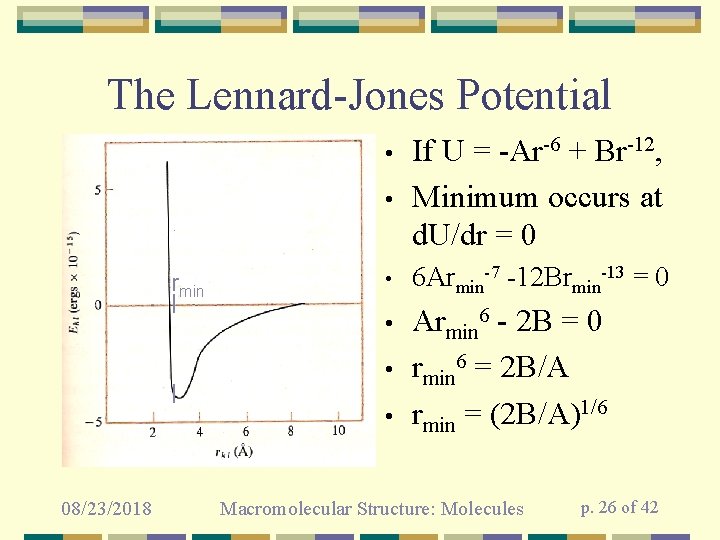
The Lennard-Jones Potential • If U = -Ar-6 + Br-12, Minimum occurs at d. U/dr = 0 • 6 Armin-7 -12 Brmin-13 = 0 • Armin 6 - 2 B = 0 rmin 6 = 2 B/A rmin = (2 B/A)1/6 • rmin • • 08/23/2018 Macromolecular Structure: Molecules p. 26 of 42

Energies of long -range interactions depend on intervening medium • • • Charges become shielded in a polar medium A vacuum is least polarizable medium, i. e. least amount of shielding Polarizability described in terms of dielectric constant D Long range interactions weaker in water than in a vacuum Dwater = 80*Dvacuum 08/23/2018 Macromolecular Structure: Molecules p. 27 of 42
Molecular interactions depend on their environment • • Cells are typically > 70% water Biochemists usually study molecules in dilute aqueous solution Easiest to treat theoretically Not always appropriate • Membrane bound proteins • Presence of "Bound Water”, e. g. groove of DNA helix • Many binding interactions driven by hydrophobic interactions 08/23/2018 Macromolecular Structure: Molecules p. 28 of 42

Structure of Water • Water has a tetrahedral structure with two hydrogens and two nonbonding electron pairs at apices • Water – water bonds are hydrogen bonds • (closest approach < sum of van der Waals radii) • Hydrogen bonds have characteristic lengths • depending on the particular hydrogen bond donor and acceptor H-O-H bond angle ~ 104º (less than tetrahedral 109. 4º): unbonded electron pairs repel more than bonds 08/23/2018 Macromolecular Structure: Molecules p. 29 of 42
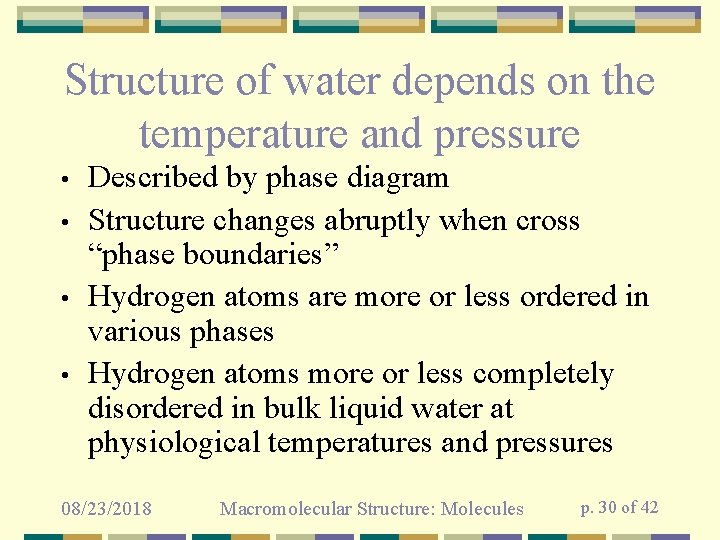
Structure of water depends on the temperature and pressure • • Described by phase diagram Structure changes abruptly when cross “phase boundaries” Hydrogen atoms are more or less ordered in various phases Hydrogen atoms more or less completely disordered in bulk liquid water at physiological temperatures and pressures 08/23/2018 Macromolecular Structure: Molecules p. 30 of 42
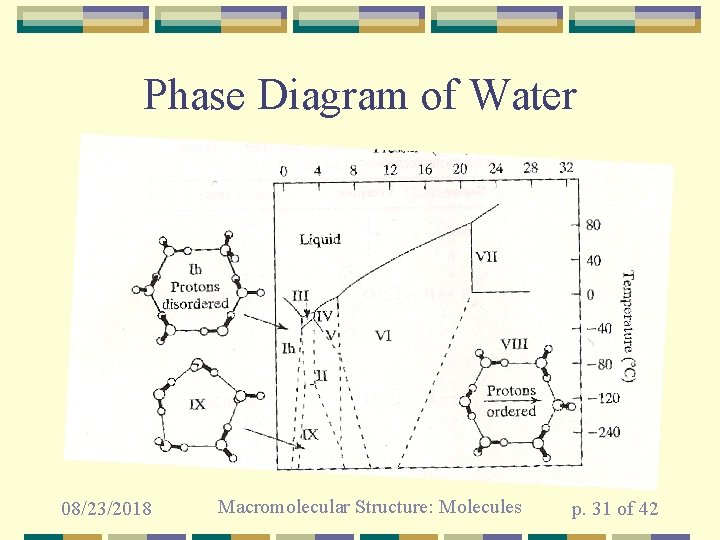
Phase Diagram of Water 08/23/2018 Macromolecular Structure: Molecules p. 31 of 42
Interaction of Molecules with Water • • • Interactions with the solvent very important for the properties of molecules in solution Molecules in solution will be surrounded by a “hydration” shell of more or less ordered water Ions overcome the decrease in entropy of the water by the enthalpy of reaction with water Hydrophobic molecules will be surrounded by highly ordered Ice-IX like structures Very low entropy, highly energetically unfavorable hence low solubility of hydrocarbons in water 08/23/2018 Macromolecular Structure: Molecules p. 32 of 42
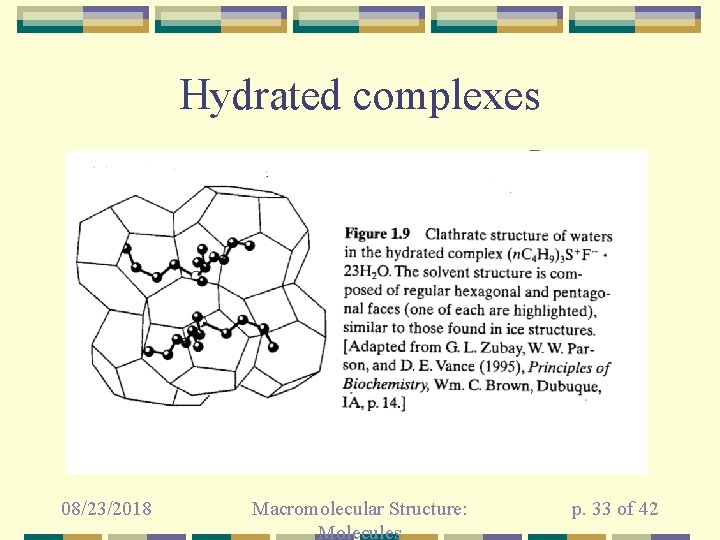
Hydrated complexes 08/23/2018 Macromolecular Structure: Molecules p. 33 of 42
Some molecules are amphipathic • • • Have both hydrophilic and hydrophobic portions e. g. phospholipids, segments of some proteins form various structures in water depending on conditions • Lipid bilayers form basis of biological membranes • Proteins and nucleic acids also amphipathic • Hydrophobic effect drives the folding of these into compact forms in water 08/23/2018 Macromolecular Structure: Molecules p. 34 of 42
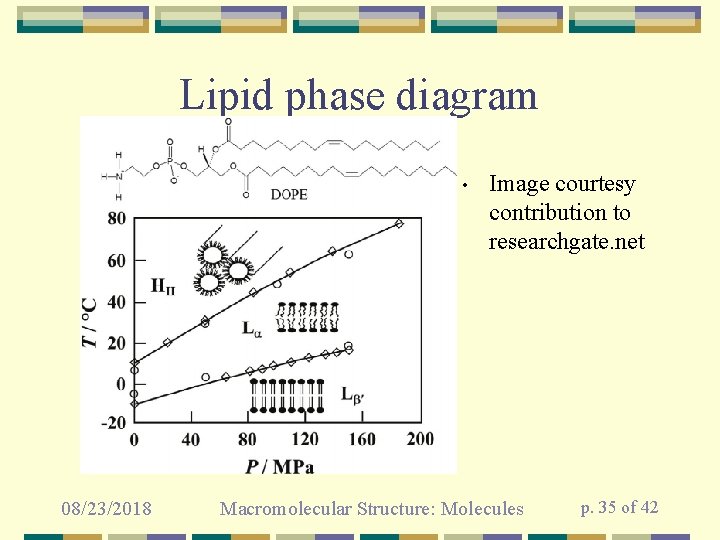
Lipid phase diagram • 08/23/2018 Image courtesy contribution to researchgate. net Macromolecular Structure: Molecules p. 35 of 42
Biological Molecules in nonaqueous environments • • • Most important case integral membrane proteins Hydrophobic groups now exposed to “solvent” Must bury hydrophilic residues or have them stick out of the membrane Energy of a charged group much higher in lipid environment than in water Consequence of dramatically lower dielectric constant which increases strength of interactions by ~40 -fold =>Very low probability you will find an unburied charge in interior of a bilayer 08/23/2018 Macromolecular Structure: Molecules p. 36 of 42
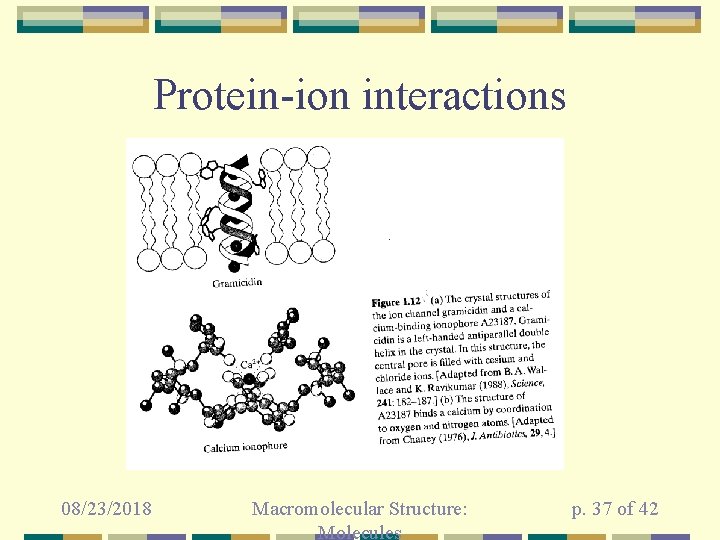
Protein-ion interactions 08/23/2018 Macromolecular Structure: Molecules p. 37 of 42
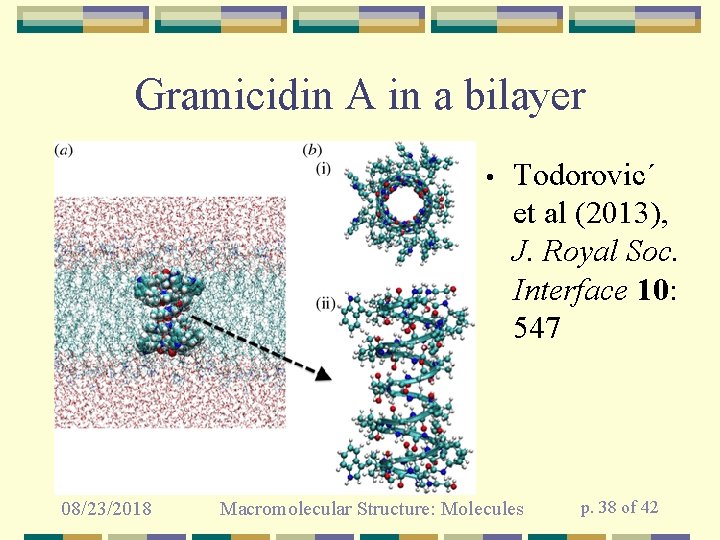
Gramicidin A in a bilayer • 08/23/2018 Todorovic´ et al (2013), J. Royal Soc. Interface 10: 547 Macromolecular Structure: Molecules p. 38 of 42

A 23187 controls 08/23/2018 2+ Ca flux Macromolecular Structure: Molecules p. 39 of 42
Bilayers create a close approximation of a 2 -D environment • • • Kinetics of reactions Rates of diffusion, etc. All very different from what you find in solution, which is an inherently threedimensional phenomenon 08/23/2018 Macromolecular Structure: Molecules p. 40 of 42
Interior of protein resembles an organic solvent • • Energetically unfavorable to bury charge Charged amino groups very different properties in interior of protein vs exterior 08/23/2018 Macromolecular Structure: Molecules p. 41 of 42
Configuration vs Conformation • Configuration refers to the arrangement of atoms around non-rotating bonds or chiral centers • Configurations can only be changed by breaking covalent bonds • cis-trans isomers • L-D stereoisomers of proteins • Molecules with the same atomic composition but different configurations may have entirely different chemistry! 08/23/2018 Macromolecular Structure: Molecules p. 42 of 42
- Slides: 42