Macromolecular Structure Middleware Open MMS An Ontology Driven











- Slides: 45

Macromolecular Structure Middleware Open. MMS An Ontology Driven Architecture http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

Overview § § The mm. CIF Ontology Open. MMS Toolkit § Macromolecular Structure (MMS) Metamodel § Parser, XML § SQL / Corba Servers and Clients § § Corba UML and the future. . . http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

How do we “Enable” Science? n n Promote well defined Macromolecular Structure (MMS) Specifications Distribution – Open Interfaces – Now: • flat files • W 3 browsing and searching – Future: • XML, SQL, CORBA http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

Why Open. MMS? n n n Allow programmers to more easily create efficient, high performance and robust applications. A Java-only toolkit with that creates XML, CORBA and Relational DB representations of the mm. CIF Macromolecular Structure Data. Source code is publicly available so users can easily modify the metamodel or create an entirely new one. http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

What Do We Mean by an Ontology Driven Architecture? What do we mean by an Ontology? A bridge between Our World of Natural Language and the World of Machines. http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

mm. CIF Dictionary and Data Files n n n Based on Ontology for Macromolecular Structure defined by the International Union of Crystallography Replaces the older 80 -Column PDB files mm. CIF Dictionary contains over 140 Category and 1600 Item definitions Open, Extensible Provides a well-defined reference standard for data distribution http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

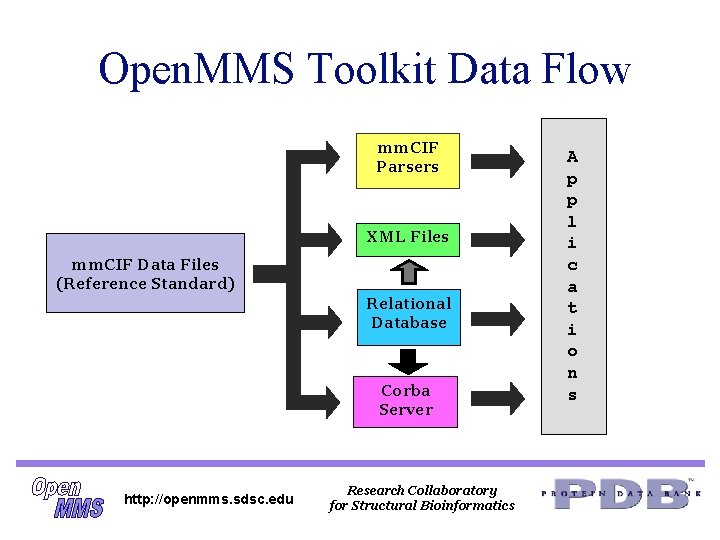
Open. MMS Toolkit Data Flow mm. CIF Parsers XML Files mm. CIF Data Files (Reference Standard) Relational Database Corba Server http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics A p p l i c a t i o n s

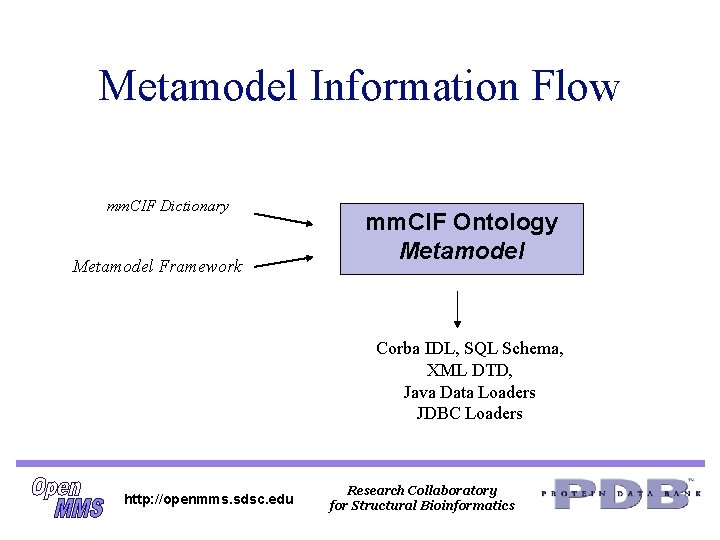
Metamodel Information Flow mm. CIF Dictionary Metamodel Framework mm. CIF Ontology Metamodel Corba IDL, SQL Schema, XML DTD, Java Data Loaders JDBC Loaders http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

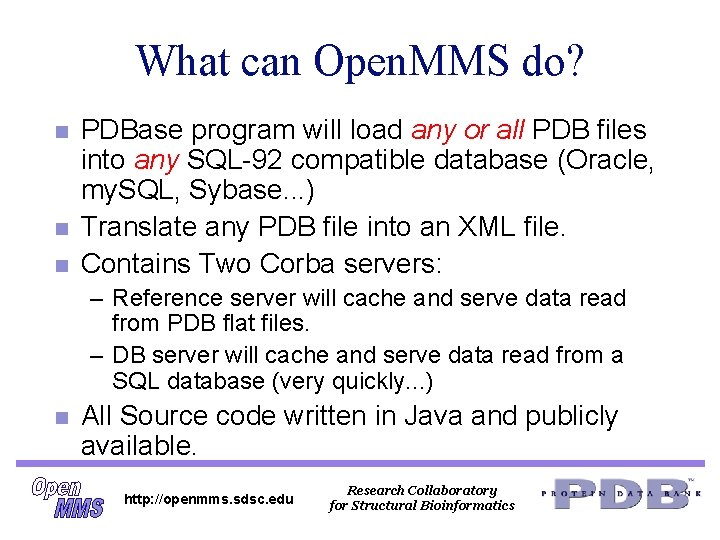
What can Open. MMS do? n n n PDBase program will load any or all PDB files into any SQL-92 compatible database (Oracle, my. SQL, Sybase. . . ) Translate any PDB file into an XML file. Contains Two Corba servers: – Reference server will cache and serve data read from PDB flat files. – DB server will cache and serve data read from a SQL database (very quickly. . . ) n All Source code written in Java and publicly available. http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

Some Advantages of Using an Ontology Driven Architecture n n n Scales to very large Ontologies More reliable and maintainable code Transfer between representations Scientific Correctness of representation Help in maintaining backward compatibility http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

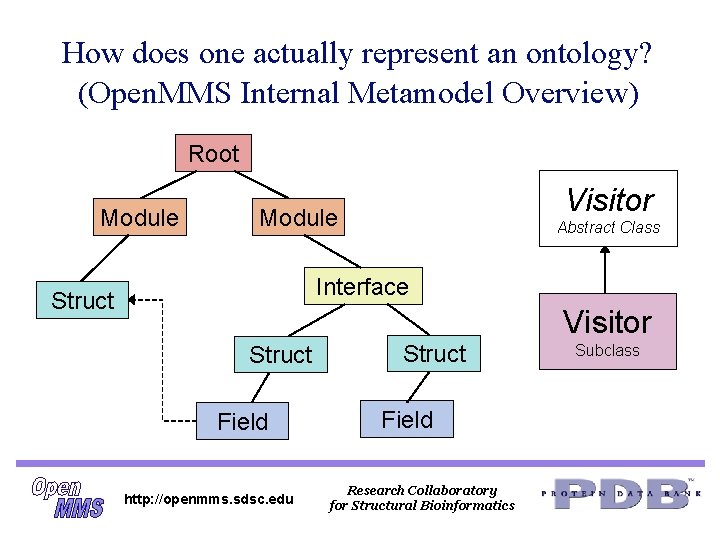
How does one actually represent an ontology? (Open. MMS Internal Metamodel Overview) Root Module Visitor Module Abstract Class Interface Struct Visitor Struct Field http: //openmms. sdsc. edu Struct Field Research Collaboratory for Structural Bioinformatics Subclass

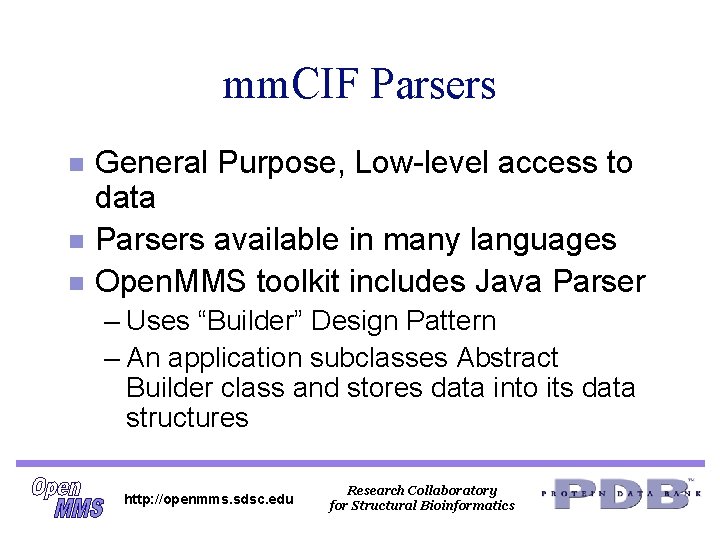
mm. CIF Parsers n n n General Purpose, Low-level access to data Parsers available in many languages Open. MMS toolkit includes Java Parser – Uses “Builder” Design Pattern – An application subclasses Abstract Builder class and stores data into its data structures http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

MMS in XML n n n Large Flat Files (open and close tags) Tables can be grouped by rows or columns XML from SQL Query – Many requests from Web browsers don’t really need or want all the data – SW available from DB Vendors and ISVs for creating XML files from SQL result sets – Smaller files load faster http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

Relational DB Expression n n SQL-92 Compatible Schemas for all the standard DB vendors Fast and Flexible Keyword searches PDBase loader allows structures to be selectively loaded Oracle Instance Tested – 14, 556 Structures – 16 GB, 88 Million Atom Records http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

A very high-level (and very-rough) classification of communication n Person-to-Person communication – email n Person-to-Machine communication – HTTP/HTML n Machine-to-Machine communication – CORBA, SQL, . NET, Soap n Not Communications -> Data Formats – XML, mm. CIF (STAR), many more … http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

What is CORBA? Common Object Request Broker Architecture Defines a family of open software interface specifications for distributed object computing. http: //www. omg. org http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

What is an Object? “A Data Structure with an Attitude” Programs = Algorithms + Data Structure Object Oriented Programming Principle: Partition the parts of algorithms with the data structures they use http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

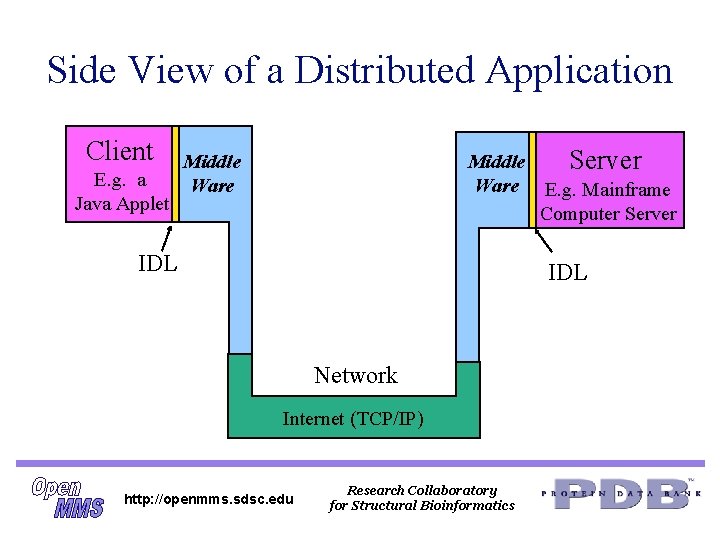
Side View of a Distributed Application Client E. g. a Java Applet Middle Server Ware E. g. Mainframe Computer Server Middle Ware IDL Network Internet (TCP/IP) http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

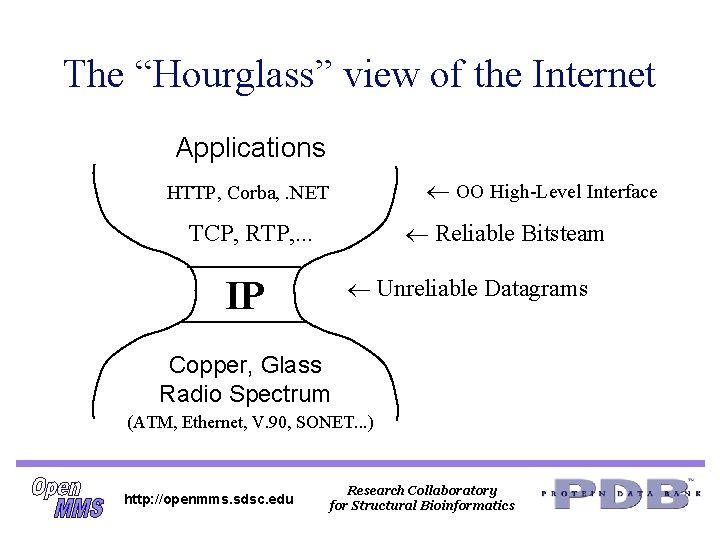
The “Hourglass” view of the Internet Applications OO High-Level Interface HTTP, Corba, . NET Reliable Bitsteam TCP, RTP, . . . Unreliable Datagrams IP Copper, Glass Radio Spectrum (ATM, Ethernet, V. 90, SONET. . . ) http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

Where is Corba? n n Inside every Java Runtime Environment. Commonly used in middle tier and backend (e. g. database) connections. Open Source and Commercial Implementations Available Usually buried deep inside the software – Difficult or impossible to tell when it is being used http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

What is Distributed Object Computing? n n Extends the benefits of object-oriented technology across process and machine boundaries to encompass entire networks. Attempts to make remote objects appear to programmers as if they were local objects in the same process. This is called location transparency. http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

Advantages of Distributed Object Computing n n n Easier (and faster) for programmers to create distributed applications Increases Reliability Increases Maintainability Increases Portability Increases Extensibility http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

The Alphabet Soup n OMG = Object Management Group Consortium of 800+ companies founded in 1989. n IDL = Interface Definition Language http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

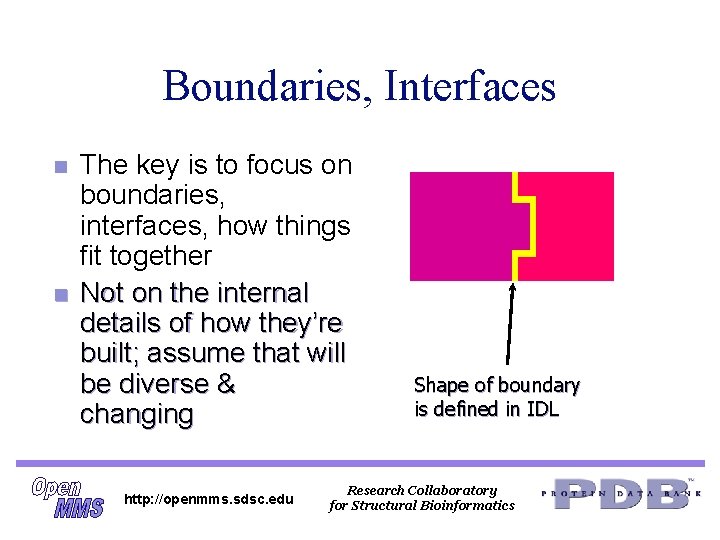
Boundaries, Interfaces n n The key is to focus on boundaries, interfaces, how things fit together Not on the internal details of how they’re built; assume that will be diverse & changing http: //openmms. sdsc. edu Shape of boundary is defined in IDL Research Collaboratory for Structural Bioinformatics

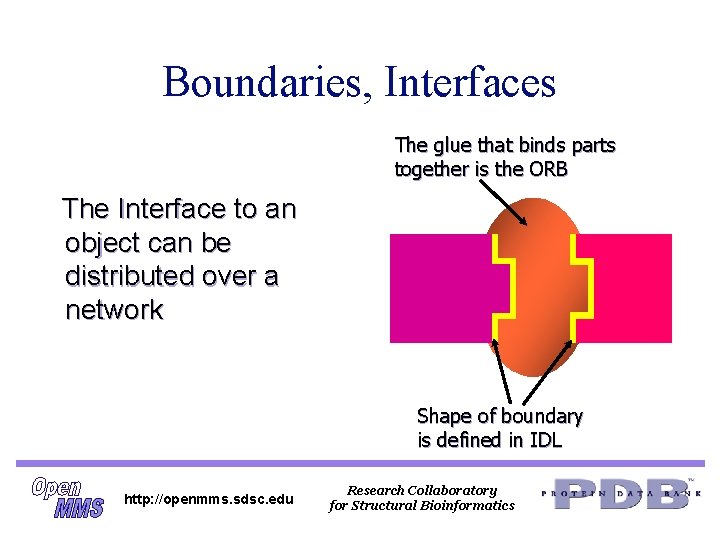
Boundaries, Interfaces The glue that binds parts together is the ORB The Interface to an object can be distributed over a network Shape of boundary is defined in IDL http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

Corba Independence n n n Open Standard for Distributed Object Oriented Design Independent of Hardware Platform Independent of Operating System Independent of Programming Language Independent of Object Location http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

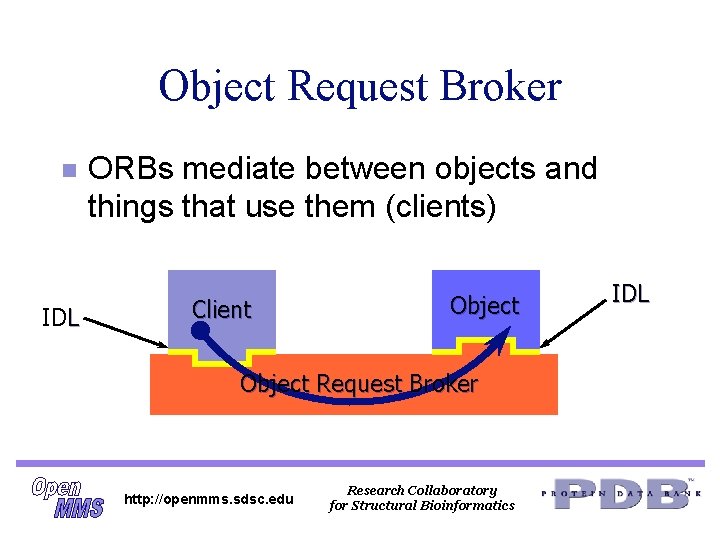
Object Request Broker n IDL ORBs mediate between objects and things that use them (clients) Client Object Request Broker http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics IDL

Terminology n IIOP – The Internet Inter-ORB Protocol, defined in the Spec as a vendor-independent, wirelevel network protocol on top of TCP/IP. This allows ORB implementations of different vendors to interoperate. http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

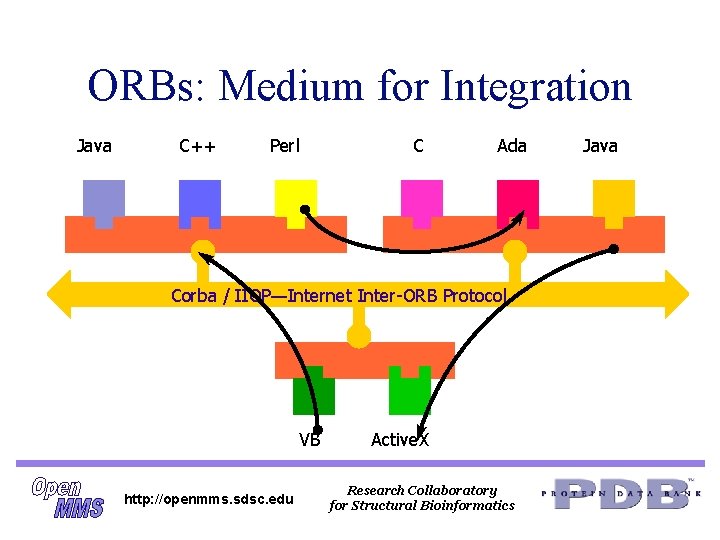
ORBs: Medium for Integration Java C++ Perl C ORB Ada ORB Corba / IIOP—Internet Inter-ORB Protocol ORB VB http: //openmms. sdsc. edu Active. X Research Collaboratory for Structural Bioinformatics Java

Corba Facilities: Industry Standards in Vertical Markets n n n Manufacturing Finance Life Sciences Research C 4 I Many others. . . http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

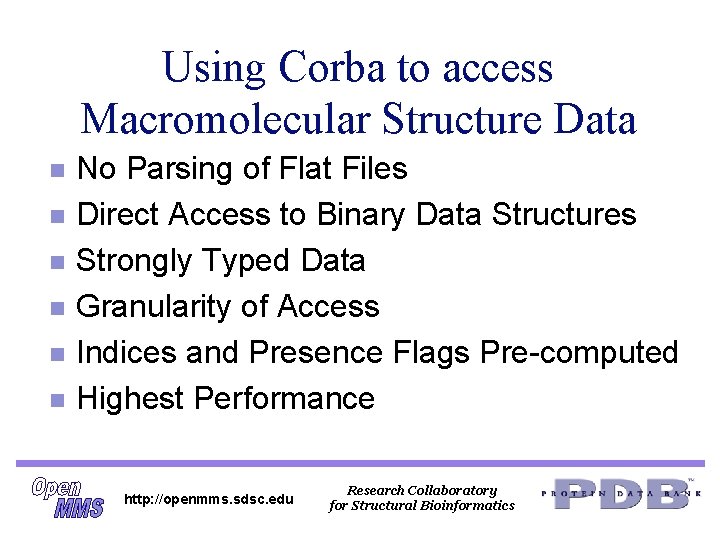
Using Corba to access Macromolecular Structure Data n n n No Parsing of Flat Files Direct Access to Binary Data Structures Strongly Typed Data Granularity of Access Indices and Presence Flags Pre-computed Highest Performance http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

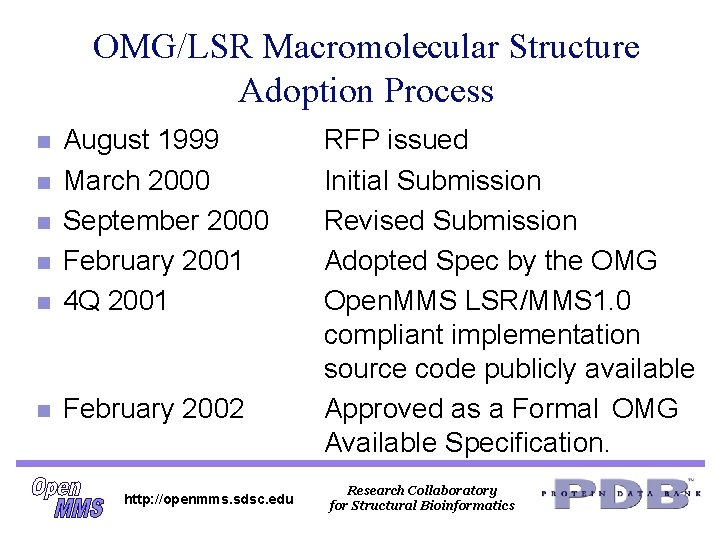
OMG/LSR Macromolecular Structure Adoption Process n August 1999 March 2000 September 2000 February 2001 4 Q 2001 n February 2002 n n http: //openmms. sdsc. edu RFP issued Initial Submission Revised Submission Adopted Spec by the OMG Open. MMS LSR/MMS 1. 0 compliant implementation source code publicly available Approved as a Formal OMG Available Specification. Research Collaboratory for Structural Bioinformatics

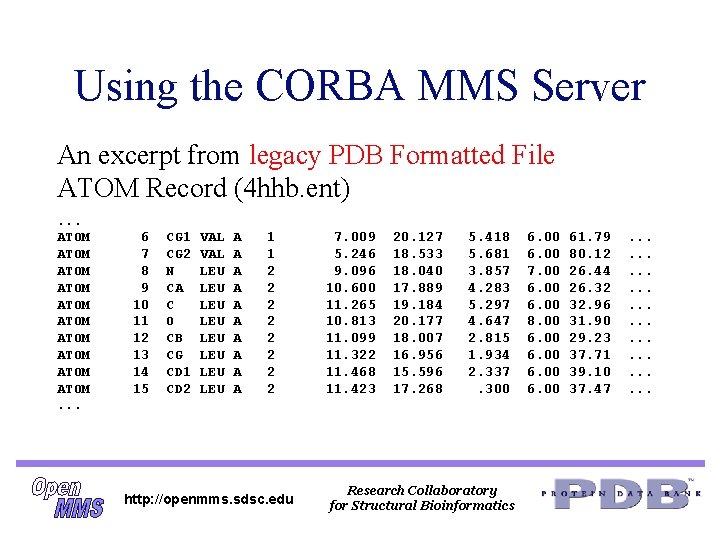
Using the CORBA MMS Server An excerpt from legacy PDB Formatted File ATOM Record (4 hhb. ent). . . ATOM ATOM ATOM. . . 6 7 8 9 10 11 12 13 14 15 CG 1 CG 2 N CA C O CB CG CD 1 CD 2 VAL LEU LEU A A A A A 1 1 2 2 2 2 http: //openmms. sdsc. edu 7. 009 5. 246 9. 096 10. 600 11. 265 10. 813 11. 099 11. 322 11. 468 11. 423 20. 127 18. 533 18. 040 17. 889 19. 184 20. 177 18. 007 16. 956 15. 596 17. 268 5. 418 5. 681 3. 857 4. 283 5. 297 4. 647 2. 815 1. 934 2. 337. 300 Research Collaboratory for Structural Bioinformatics 6. 00 7. 00 6. 00 8. 00 61. 79 80. 12 26. 44 26. 32 32. 96 31. 90 29. 23 37. 71 39. 10 37. 47 . . . .

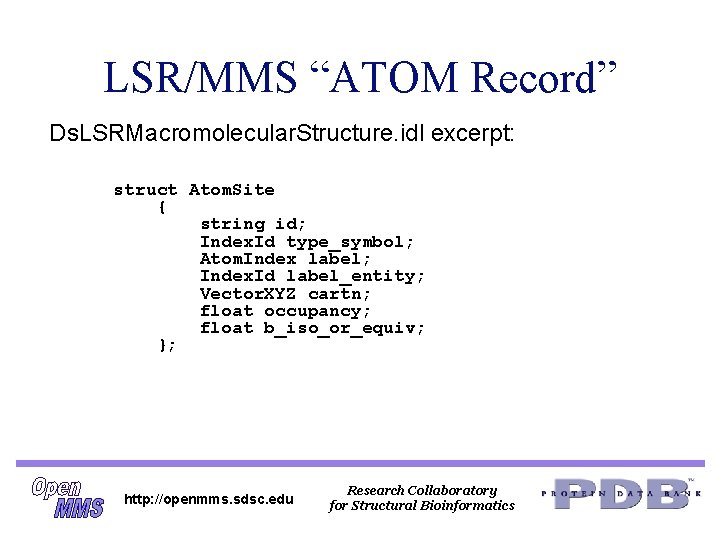
LSR/MMS “ATOM Record” Ds. LSRMacromolecular. Structure. idl excerpt: struct Atom. Site { string id; Index. Id type_symbol; Atom. Index label; Index. Id label_entity; Vector. XYZ cartn; float occupancy; float b_iso_or_equiv; }; http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

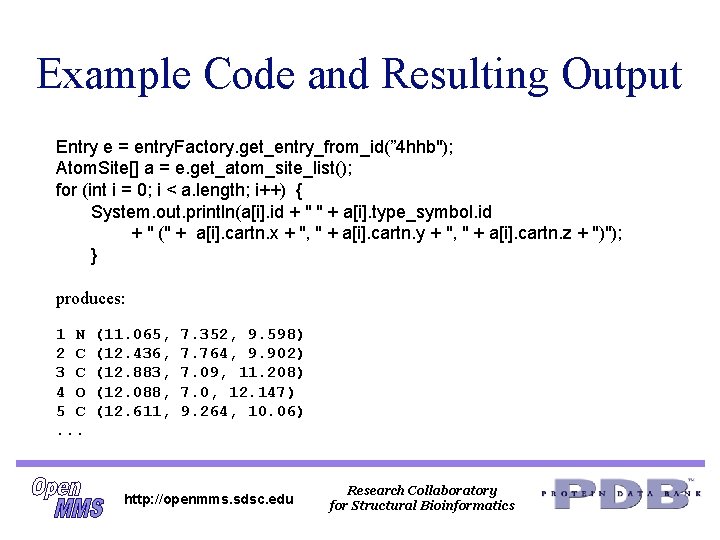
Example Code and Resulting Output Entry e = entry. Factory. get_entry_from_id(” 4 hhb"); Atom. Site[] a = e. get_atom_site_list(); for (int i = 0; i < a. length; i++) { System. out. println(a[i]. id + " " + a[i]. type_symbol. id + " (" + a[i]. cartn. x + ", " + a[i]. cartn. y + ", " + a[i]. cartn. z + ")"); } produces: 1 N 2 C 3 C 4 O 5 C. . . (11. 065, (12. 436, (12. 883, (12. 088, (12. 611, 7. 352, 9. 598) 7. 764, 9. 902) 7. 09, 11. 208) 7. 0, 12. 147) 9. 264, 10. 06) http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

What are the alternatives to Corba? n TCP/IP Sockets - Byte stream n DCOM, COM++, OLE, . NET (Microsoft Only) – DCOM Corba Bridges are available from several vendors n SOAP (Simple Object Access Protocol) – XML Based http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

Unified Modeling Language – UML What do all those arrows and boxes Mean? n n n Schematic Language for Defining SW Graphics Representations UML = Things, Relations and Diagrams 9 types of Diagrams The most commonly used diagram is the “Class Diagram” http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

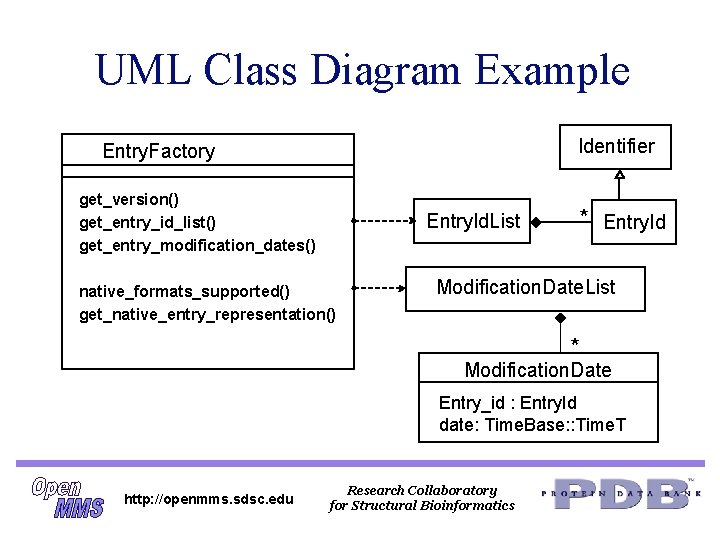
UML Class Diagram Example Identifier Entry. Factory get_version() get_entry_id_list() get_entry_modification_dates() Entry. Id. List native_formats_supported() get_native_entry_representation() * Entry. Id Modification. Date. List * Modification. Date Entry_id : Entry. Id date: Time. Base: : Time. T http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

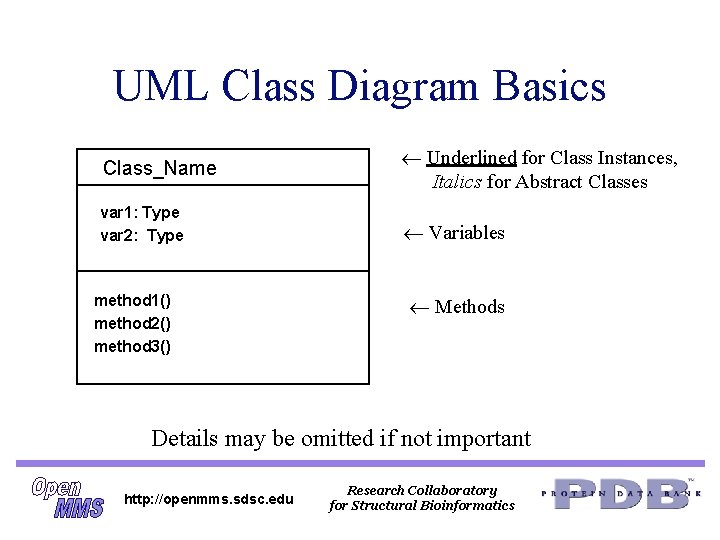
UML Class Diagram Basics Class_Name Underlined for Class Instances, Italics for Abstract Classes var 1: Type var 2: Type Variables method 1() method 2() method 3() Methods Details may be omitted if not important http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

UML Relationships Dependency 0. . 1 * Association Generalization (Inheritance) * http: //openmms. sdsc. edu Aggregation Research Collaboratory for Structural Bioinformatics

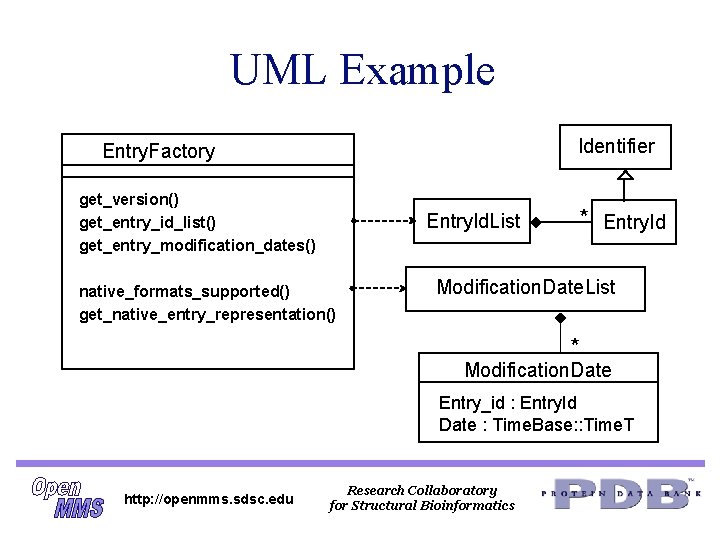
UML Example Identifier Entry. Factory get_version() get_entry_id_list() get_entry_modification_dates() Entry. Id. List native_formats_supported() get_native_entry_representation() * Entry. Id Modification. Date. List * Modification. Date Entry_id : Entry. Id Date : Time. Base: : Time. T http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

XMI: XML Metadata Interchange n n UML is a graphical representation; need some way to exchange UML models between applications XMI is used to store and transmit UML models XML based Defines XML tags for classes, relationships between classes etc. http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

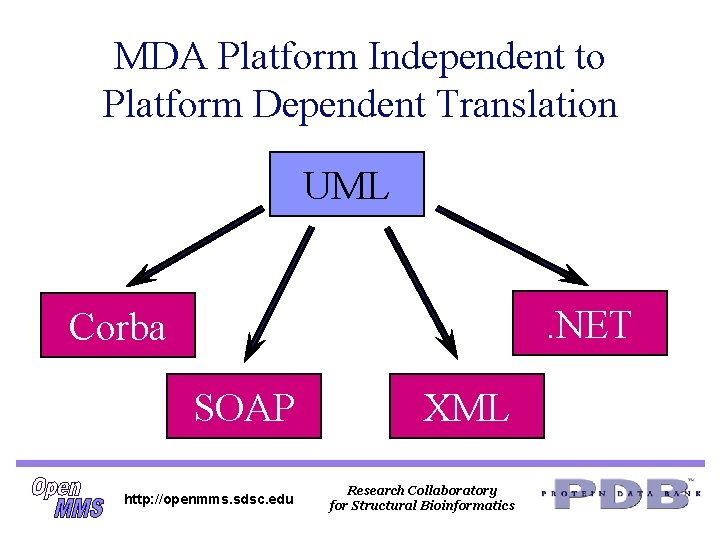
OMG MDA n n n Platform Independent Models (PIMs) that define the interface are defined in UML The PIMs are translated to Platform Specific Models (PSMs) such as Corba, SOAP, . NET or XML Schemas The Corba servers and clients may be the same, but now the interface is defined in UML and the IDL is then generated from the UML http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

MDA Platform Independent to Platform Dependent Translation UML. NET Corba SOAP XML http: //openmms. sdsc. edu Research Collaboratory for Structural Bioinformatics

Thanks and Acknowledgments Phil Bourne John Westbrook David Benton http: //openmms. sdsc. edu Karl Konnerth Lynn Ten. Eyck Research Collaboratory for Structural Bioinformatics