Macromolecular Chemistry Lecture 8 Chemistry 367 L392 N
![Molecular Weight from [h] Mark-Houwink-Kuhn-Sakurada equation [h ] = K M a a≈0 a Molecular Weight from [h] Mark-Houwink-Kuhn-Sakurada equation [h ] = K M a a≈0 a](https://slidetodoc.com/presentation_image_h2/967cada8dcc6b48761c7e61cf180794d/image-4.jpg)



- Slides: 31

Macromolecular Chemistry Lecture 8 Chemistry 367 L/392 N

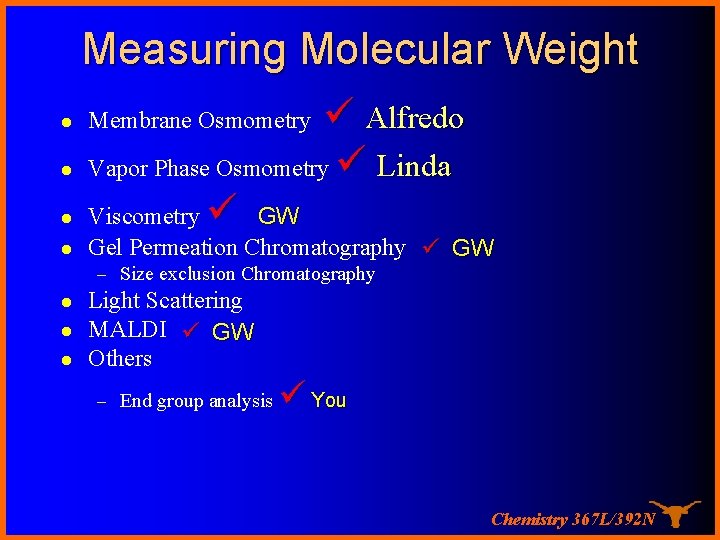
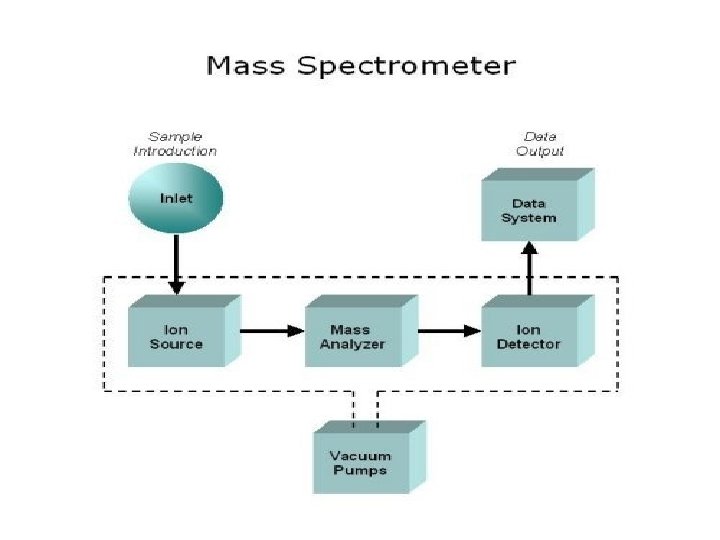
Measuring Molecular Weight l Alfredo Vapor Phase Osmometry Linda Viscometry GW l Gel Permeation Chromatography GW l l Membrane Osmometry – Size exclusion Chromatography l l l Light Scattering MALDI GW Others – End group analysis You Chemistry 367 L/392 N

Chemistry 367 L/392 N
![Molecular Weight from h MarkHouwinkKuhnSakurada equation h K M a a0 a Molecular Weight from [h] Mark-Houwink-Kuhn-Sakurada equation [h ] = K M a a≈0 a](https://slidetodoc.com/presentation_image_h2/967cada8dcc6b48761c7e61cf180794d/image-4.jpg)
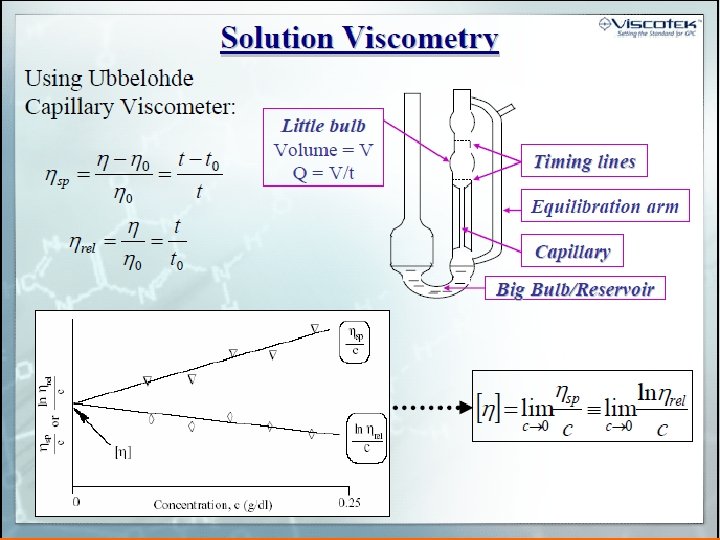
Molecular Weight from [h] Mark-Houwink-Kuhn-Sakurada equation [h ] = K M a a≈0 a = 0. 5 -0. 8 a = 1. 8 Chemistry 367 L/392 N

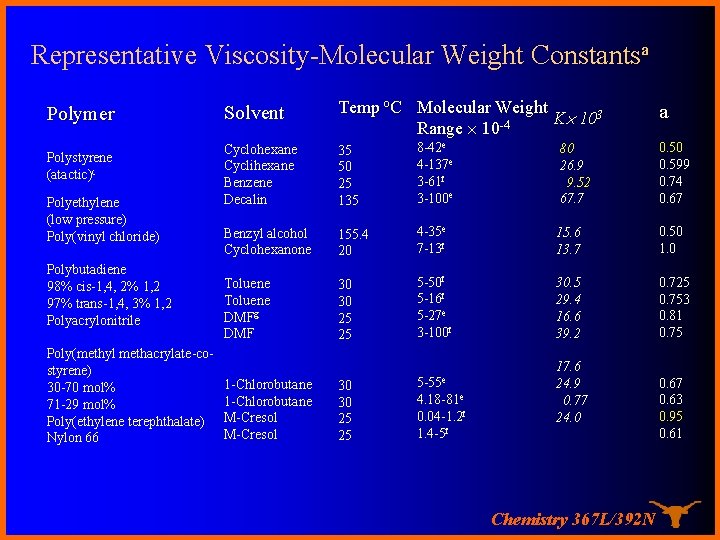
Representative Viscosity-Molecular Weight Constantsa Polymer Polystyrene (atactic)c Polyethylene (low pressure) Poly(vinyl chloride) Polybutadiene 98% cis-1, 4, 2% 1, 2 97% trans-1, 4, 3% 1, 2 Polyacrylonitrile Poly(methyl methacrylate-costyrene) 30 -70 mol% 71 -29 mol% Poly(ethylene terephthalate) Nylon 66 Solvent Temp o. C Molecular Weight K 103 -4 Range 10 Cyclohexane Cyclihexane Benzene Decalin 35 50 25 135 8 -42 e 4 -137 e 3 -61 f 3 -100 e 80 26. 9 9. 52 67. 7 0. 50 0. 599 0. 74 0. 67 Benzyl alcohol Cyclohexanone 155. 4 20 4 -35 e 7 -13 f 15. 6 13. 7 0. 50 1. 0 Toluene DMFg DMF 30 30 25 25 5 -50 f 5 -16 f 5 -27 e 3 -100 f 30. 5 29. 4 16. 6 39. 2 0. 725 0. 753 0. 81 0. 75 30 30 25 25 5 -55 e 1 -Chlorobutane M-Cresol 4. 18 -81 e 0. 04 -1. 2 f 1. 4 -5 f 17. 6 24. 9 0. 77 24. 0 Chemistry 367 L/392 N a 0. 67 0. 63 0. 95 0. 61

Mass Spectroscopy MALDI Chemistry 367 L/392 N

Chemistry 367 L/392 N

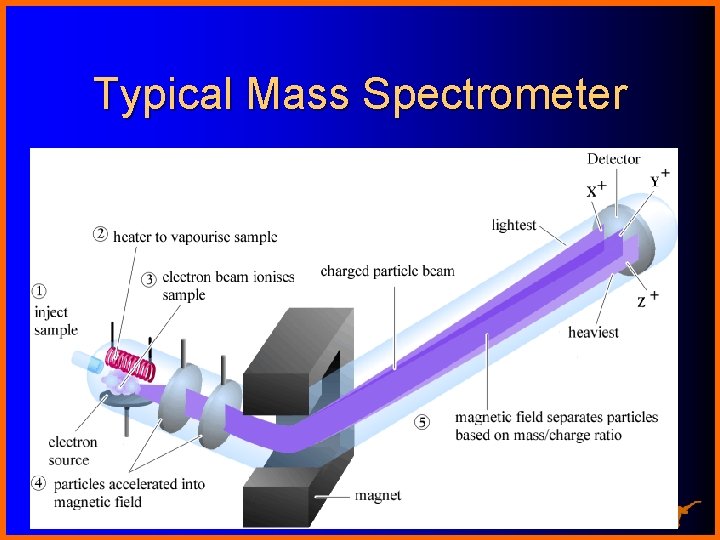
Typical Mass Spectrometer Chemistry 367 L/392 N

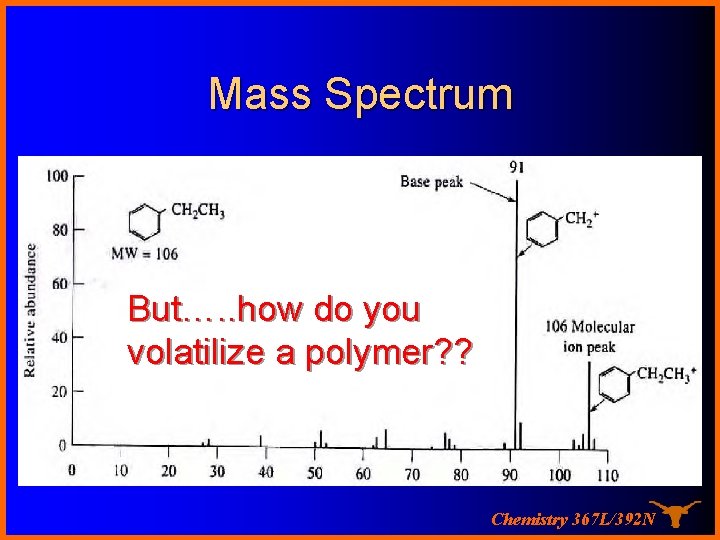
Mass Spectrum But…. . how do you volatilize a polymer? ? Chemistry 367 L/392 N

Answer: By ESI & MALDI !! Chemistry 367 L/392 N

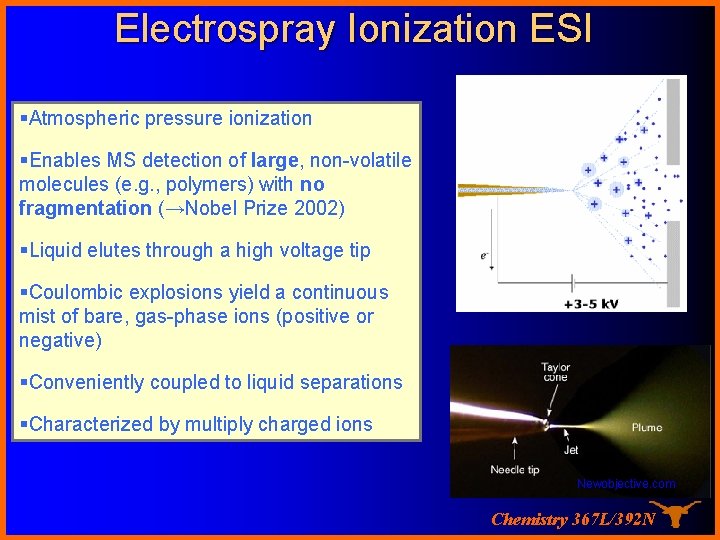
Electrospray Ionization ESI §Atmospheric pressure ionization §Enables MS detection of large, non-volatile molecules (e. g. , polymers) with no fragmentation (→Nobel Prize 2002) §Liquid elutes through a high voltage tip §Coulombic explosions yield a continuous mist of bare, gas-phase ions (positive or negative) §Conveniently coupled to liquid separations §Characterized by multiply charged ions Newobjective. com Chemistry 367 L/392 N

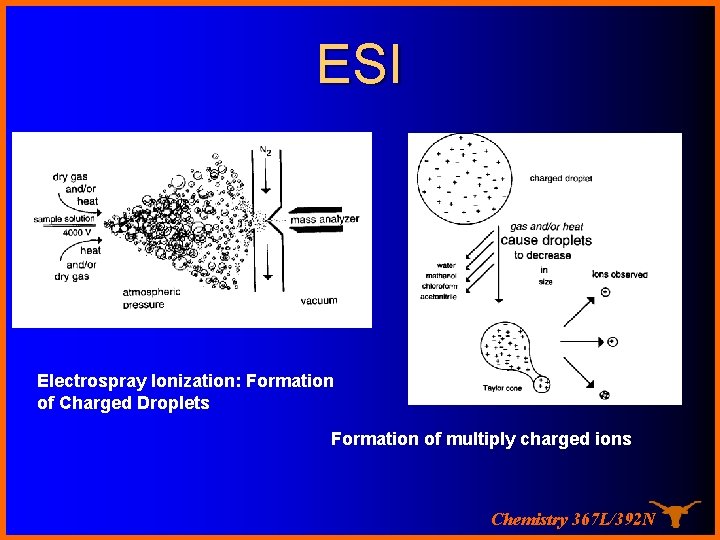
ESI Electrospray Ionization: Formation of Charged Droplets Formation of multiply charged ions Chemistry 367 L/392 N

Electrospray Chemistry 367 L/392 N

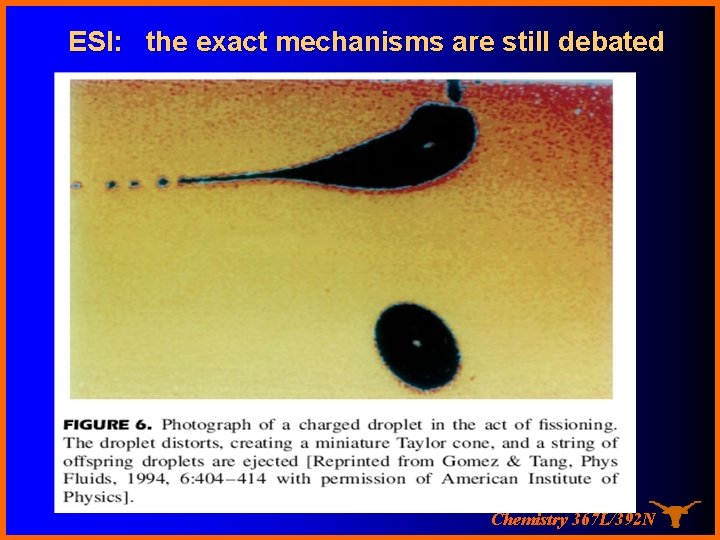
ESI: the exact mechanisms are still debated Chemistry 367 L/392 N

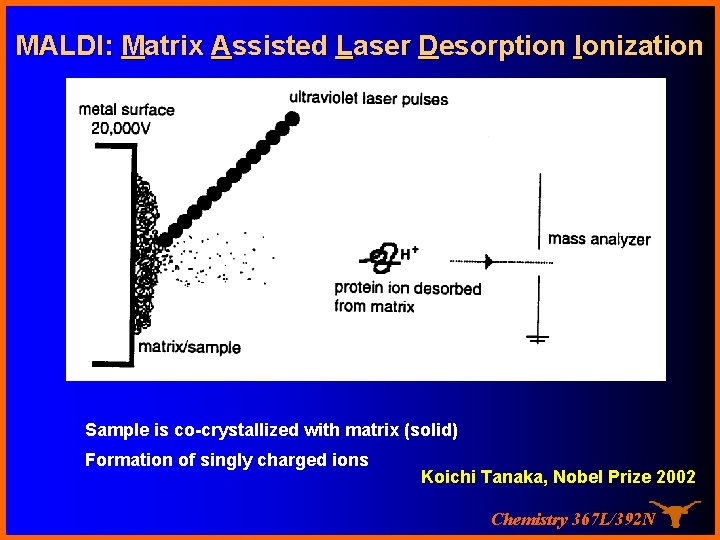
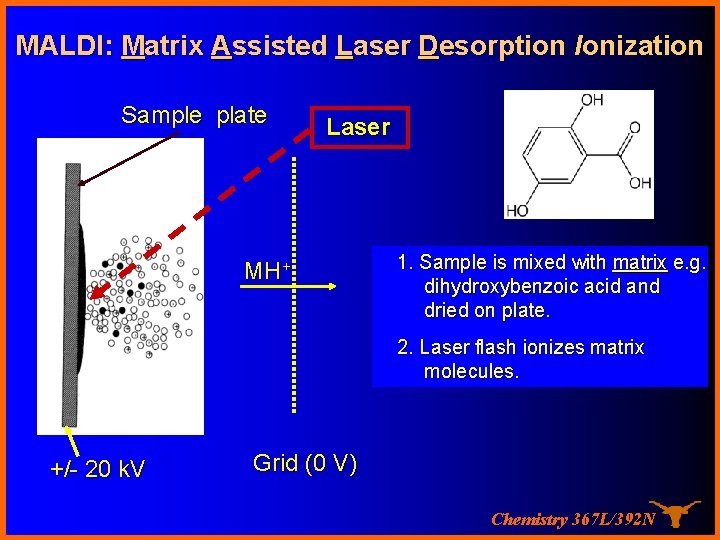
MALDI: Matrix Assisted Laser Desorption Ionization Sample is co-crystallized with matrix (solid) Formation of singly charged ions Koichi Tanaka, Nobel Prize 2002 Chemistry 367 L/392 N

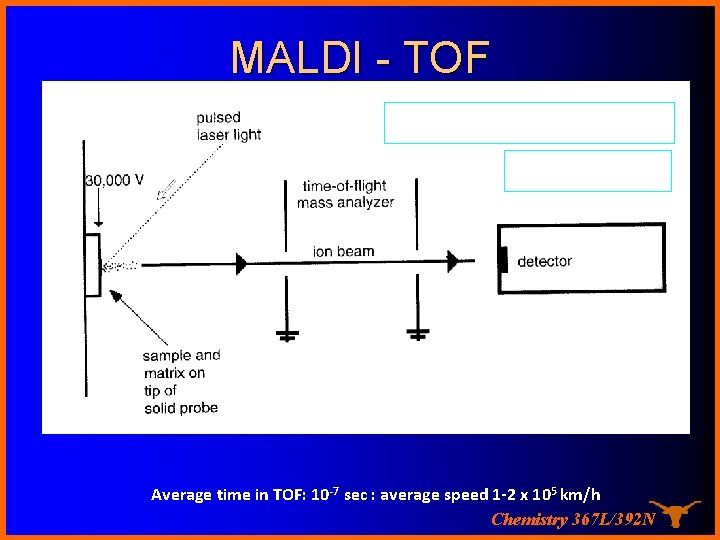
MALDI - TOF MALDI Time-of-flight MALDI TOF Drift region (D) Average time in TOF: 10 -7 sec : average speed 1 -2 x 105 km/h Chemistry 367 L/392 N

MALDI: Matrix Assisted Laser Desorption Ionization Sample plate Laser hn MH+ 1. Sample is mixed with matrix e. g. dihydroxybenzoic acid and dried on plate. 2. Laser flash ionizes matrix molecules. +/- 20 k. V Grid (0 V) Chemistry 367 L/392 N

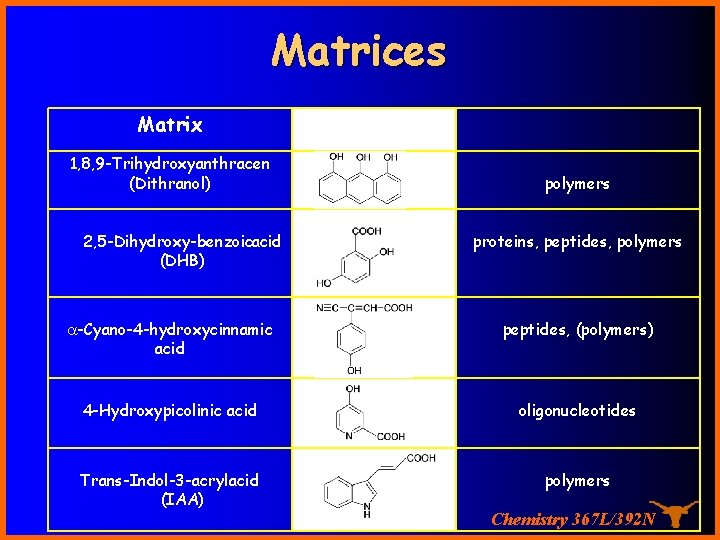
Matrices Matrix 1, 8, 9 -Trihydroxyanthracen (Dithranol) 2, 5 -Dihydroxy benzoicacid (DHB) polymers proteins, peptides, polymers -Cyano-4 -hydroxycinnamic acid peptides, (polymers) 4 -Hydroxypicolinic acid oligonucleotides Trans-Indol-3 -acrylacid (IAA) polymers Chemistry 367 L/392 N

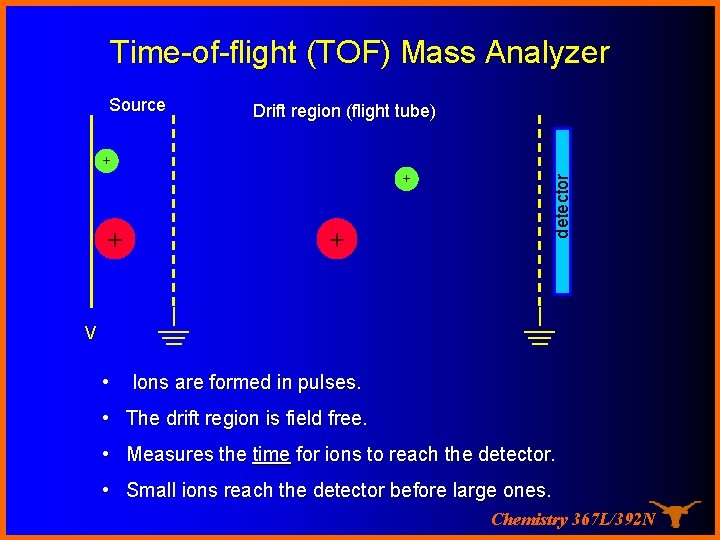
Time-of-flight (TOF) Mass Analyzer Drift region (flight tube) + + detector Source V • Ions are formed in pulses. • The drift region is field free. • Measures the time for ions to reach the detector. • Small ions reach the detector before large ones. Chemistry 367 L/392 N

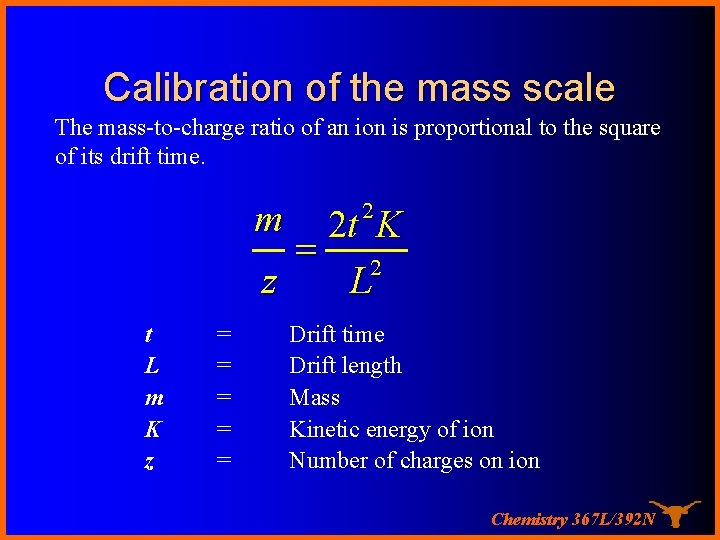
Calibration of the mass scale The mass-to-charge ratio of an ion is proportional to the square of its drift time. m 2 2 t K = 2 z L t L m K z = = = Drift time Drift length Mass Kinetic energy of ion Number of charges on ion Chemistry 367 L/392 N

Chemistry 367 L/392 N

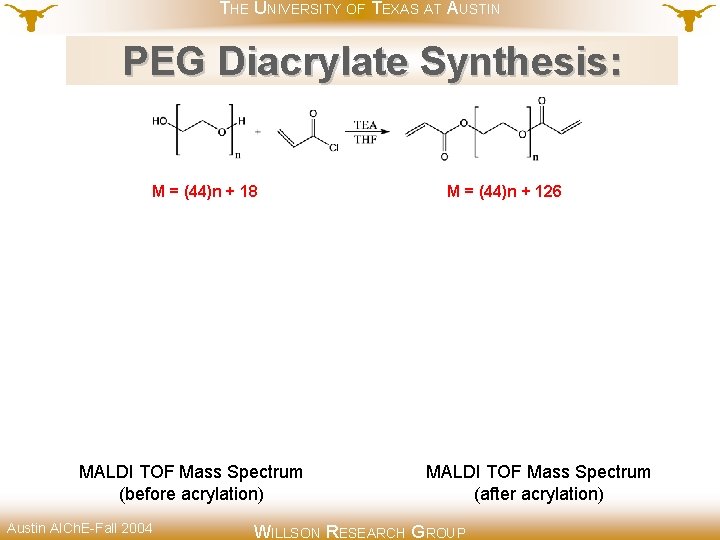
THE UNIVERSITY OF TEXAS AT AUSTIN PEG Diacrylate Synthesis: M = (44)n + 18 MALDI TOF Mass Spectrum (before acrylation) Austin AICh. E-Fall 2004 M = (44)n + 126 MALDI TOF Mass Spectrum (after acrylation) WILLSON RESEARCH GROUP

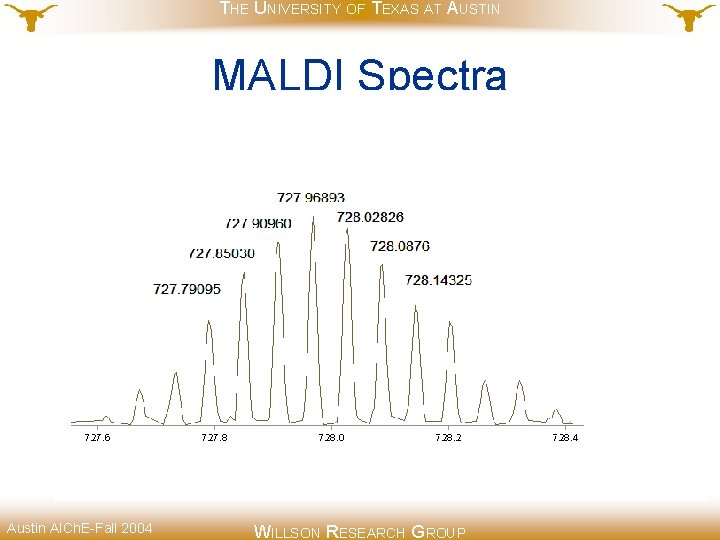
THE UNIVERSITY OF TEXAS AT AUSTIN MALDI Spectra Austin AICh. E-Fall 2004 WILLSON RESEARCH GROUP

Monoisotopic mass When the isotopes are clearly resolved the monoisotopic mass is used as it is the most accurate measurement. Chemistry 367 L/392 N

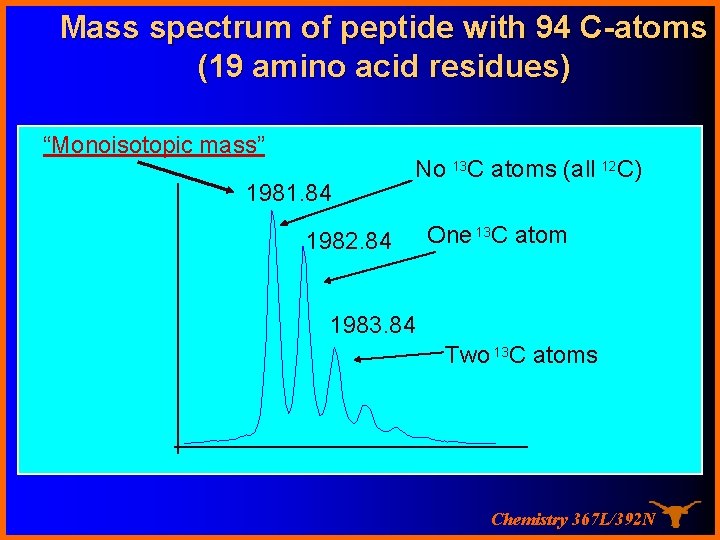
Mass spectrum of peptide with 94 C-atoms (19 amino acid residues) “Monoisotopic mass” 1981. 84 No 13 C atoms (all 12 C) 1982. 84 One 13 C atom 1983. 84 Two 13 C atoms Chemistry 367 L/392 N

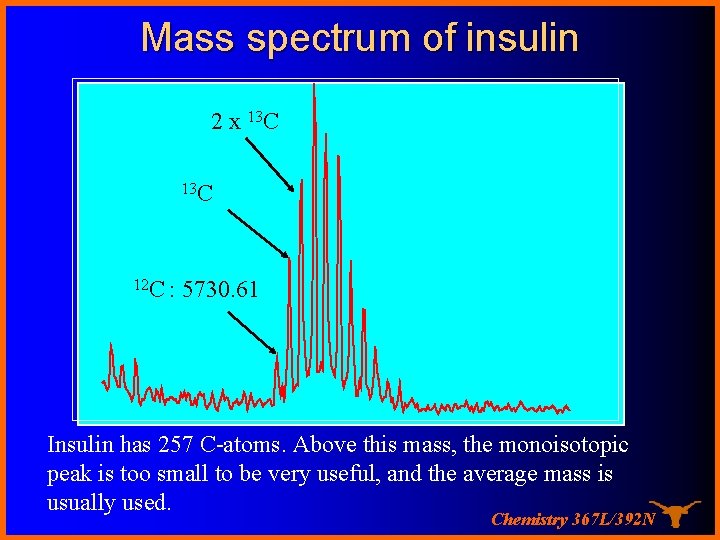
Mass spectrum of insulin 2 x 13 C 12 C : 5730. 61 Insulin has 257 C-atoms. Above this mass, the monoisotopic peak is too small to be very useful, and the average mass is usually used. Chemistry 367 L/392 N

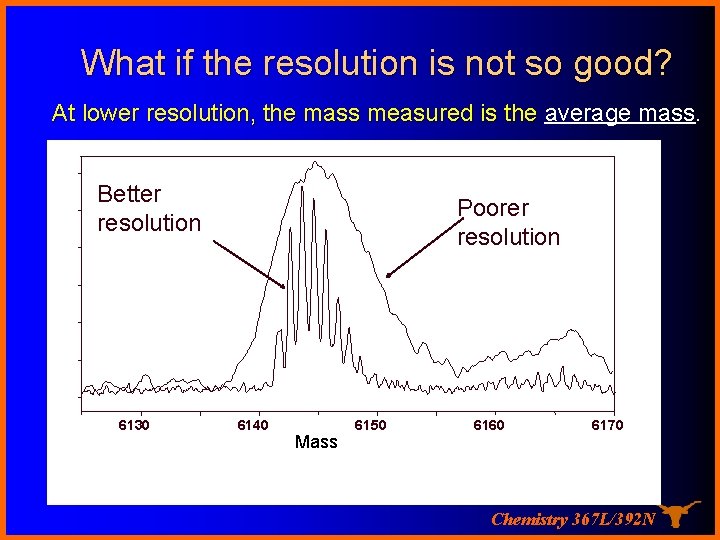
What if the resolution is not so good? At lower resolution, the mass measured is the average mass. Better resolution 6130 Poorer resolution 6140 Mass 6150 6160 6170 Chemistry 367 L/392 N

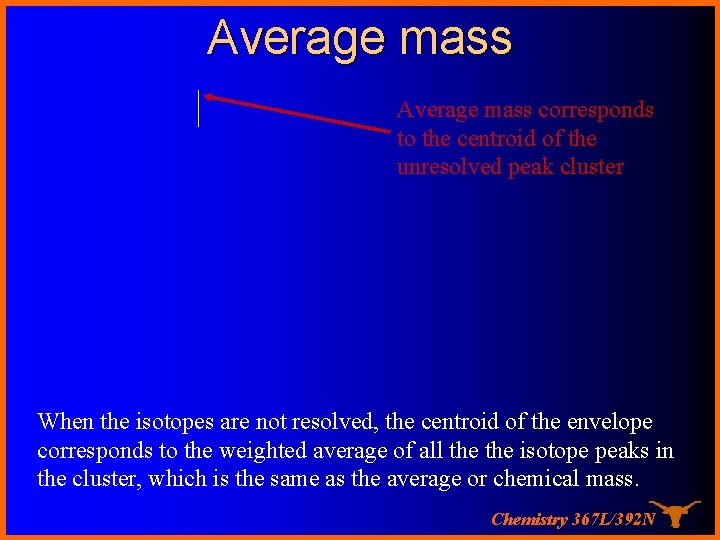
Average mass corresponds to the centroid of the unresolved peak cluster When the isotopes are not resolved, the centroid of the envelope corresponds to the weighted average of all the isotope peaks in the cluster, which is the same as the average or chemical mass. Chemistry 367 L/392 N

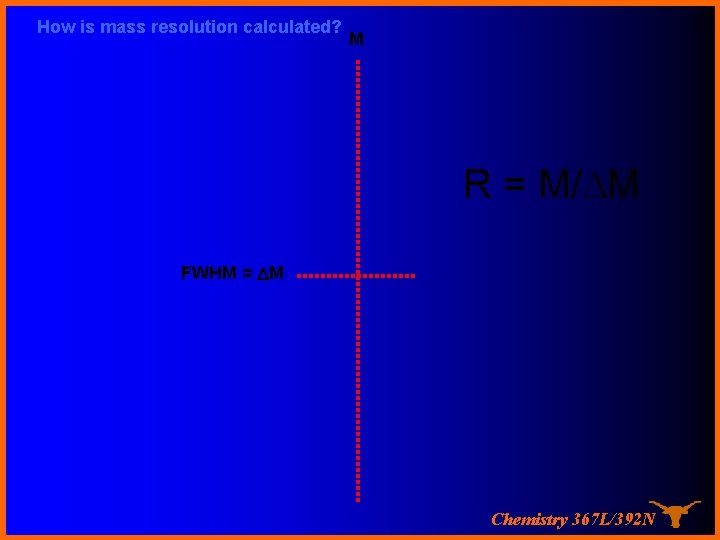
How is mass resolution calculated? M R = M/DM FWHM = DM Chemistry 367 L/392 N

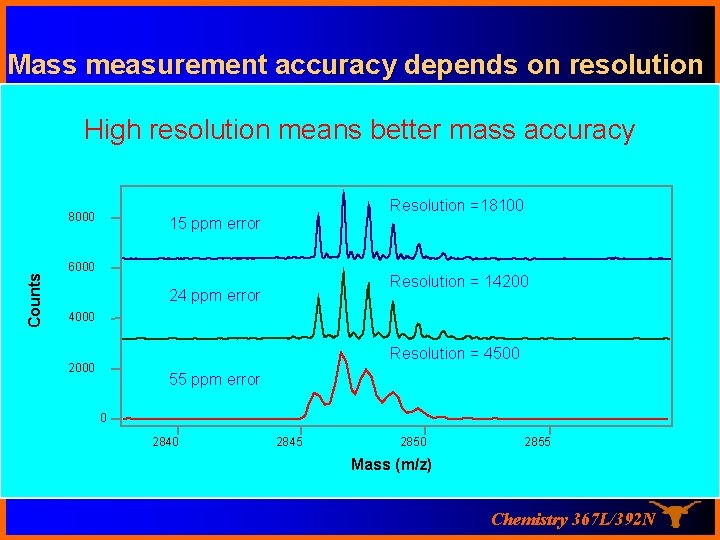
Mass measurement accuracy depends on resolution High resolution means better mass accuracy Counts 8000 Resolution =18100 15 ppm error 6000 Resolution = 14200 24 ppm error 4000 Resolution = 4500 2000 55 ppm error 0 2845 2850 2855 Mass (m/z) Chemistry 367 L/392 N

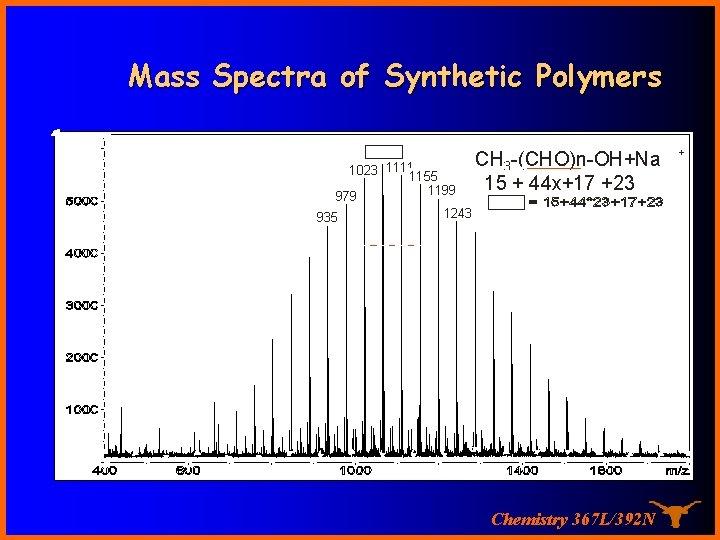
Mass Spectra of Synthetic Polymers 1067 1023 11111155 979 935 1199 CH 3 -(CHO)n-OH+Na 15 + 44 x+17 +23 1243 Chemistry 367 L/392 N +
 Cmput 365
Cmput 365 What is direct instruction strategy
What is direct instruction strategy Poder judicial
Poder judicial R v allen (1872) lr 1 ccr 367
R v allen (1872) lr 1 ccr 367 Cs 367
Cs 367 R v allen (1872) lr 1 ccr 367
R v allen (1872) lr 1 ccr 367 Cs367
Cs367 Mischief rule
Mischief rule Rounding numbers to the nearest hundred thousand
Rounding numbers to the nearest hundred thousand 01:640:244 lecture notes - lecture 15: plat, idah, farad
01:640:244 lecture notes - lecture 15: plat, idah, farad Advanced inorganic chemistry lecture notes
Advanced inorganic chemistry lecture notes Lightning elves
Lightning elves Ib chemistry functional groups
Ib chemistry functional groups Inorganic vs organic chemistry
Inorganic vs organic chemistry Dolls eyes exam
Dolls eyes exam Drawing lecture
Drawing lecture Trois femmes puissantes lecture cursive
Trois femmes puissantes lecture cursive Transformer lecture
Transformer lecture Lecture is derived from
Lecture is derived from Magnetism
Magnetism Graph neural network lecture
Graph neural network lecture Hamza kashgari
Hamza kashgari Make up lecture
Make up lecture Nlp lecture notes
Nlp lecture notes Significance of research methodology
Significance of research methodology Motivation lecture
Motivation lecture Physical science lecture notes
Physical science lecture notes Philosopher giving a lecture on the orrery
Philosopher giving a lecture on the orrery Data visualization lecture
Data visualization lecture Lecture etymology
Lecture etymology Voix haute
Voix haute Operating systems lecture notes
Operating systems lecture notes