Machine Learning as Applied to Structural Bioinformatics Results
Machine Learning as Applied to Structural Bioinformatics: Results and Challenges Philip E. Bourne University of California San Diego pbourne@ucsd. edu 10/31/2021 DIMACS - Machine Learning in Bioinformatics 1

The Current Situation • Structure contributes greatly to our understanding of living systems • We are locked into thinking about structure in specific ways which limits our view – All too often we consider structure as a static entity – The view at left is not how another protein or a small molecule ligand sees PKA • We are still not very good at certain problems … 10/31/2021 DIMACS - Machine Learning in Bioinformatics 2
Example Unsolved Problems that Machine Learning Can Address • Predicting flexibility and disorder in protein structure • Predicting sites of protein-protein and protein-ligand interaction • Predicting protein function • Defining domain boundaries from sequence • Predicting secondary, tertiary and quaternary structure • Predicting what will crystallize 10/31/2021 DIMACS - Machine Learning in Bioinformatics 3
Example Unsolved Problems that Machine Learning Can Address • Predicting flexibility and disorder in protein structure • Predicting sites of protein-protein and protein-ligand interaction • Predicting protein function • Defining domain boundaries from sequence • Predicting secondary, tertiary and quaternary structure • Predicting what will crystallize * Will talk about this * Will offer as a challenge 10/31/2021 DIMACS - Machine Learning in Bioinformatics 4
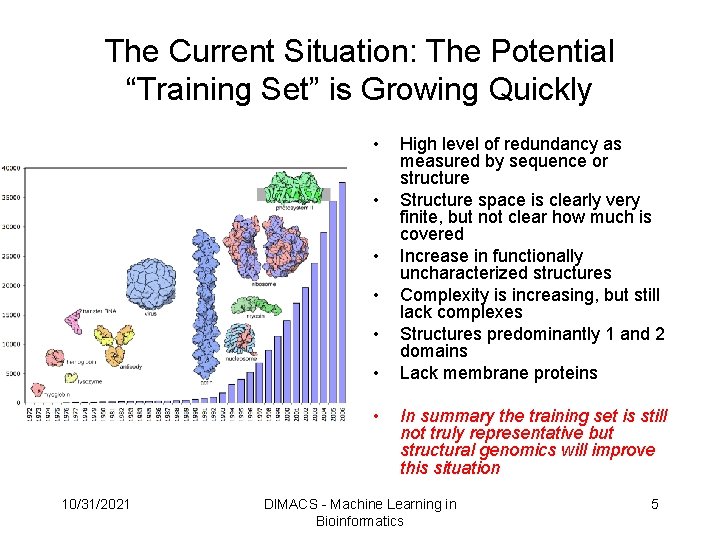
The Current Situation: The Potential “Training Set” is Growing Quickly • • 10/31/2021 High level of redundancy as measured by sequence or structure Structure space is clearly very finite, but not clear how much is covered Increase in functionally uncharacterized structures Complexity is increasing, but still lack complexes Structures predominantly 1 and 2 domains Lack membrane proteins In summary the training set is still not truly representative but structural genomics will improve this situation DIMACS - Machine Learning in Bioinformatics 5
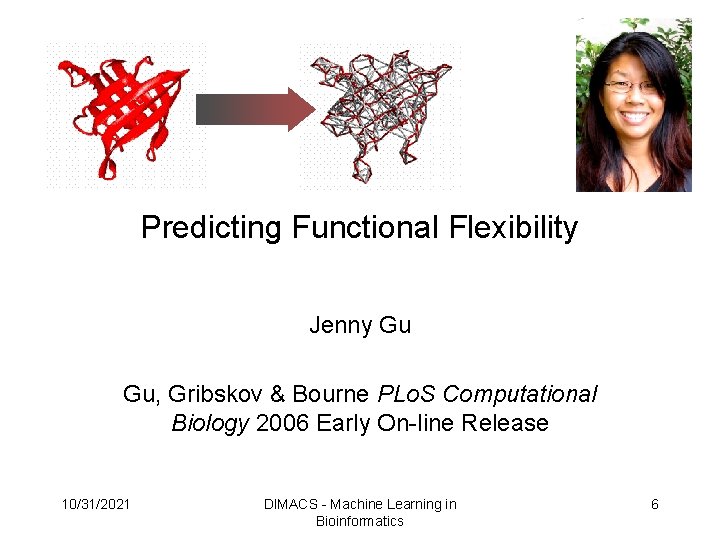
Predicting Functional Flexibility Jenny Gu Gu, Gribskov & Bourne PLo. S Computational Biology 2006 Early On-line Release 10/31/2021 DIMACS - Machine Learning in Bioinformatics 6
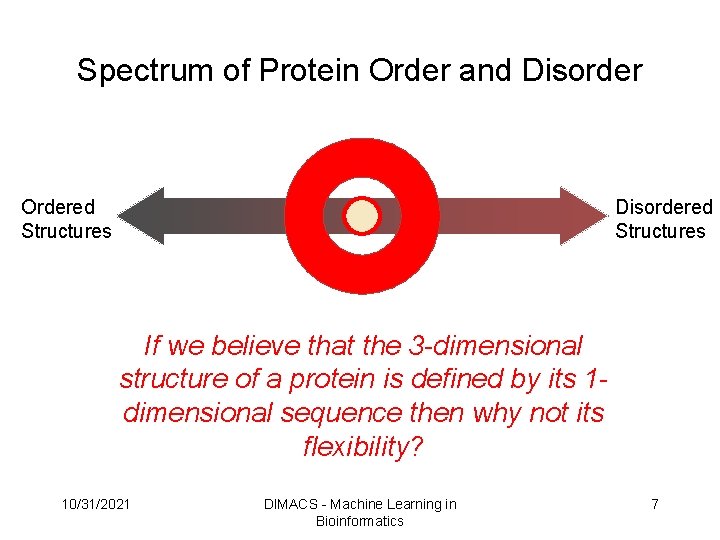
Spectrum of Protein Order and Disorder Ordered Structures Disordered Structures If we believe that the 3 -dimensional structure of a protein is defined by its 1 dimensional sequence then why not its flexibility? 10/31/2021 DIMACS - Machine Learning in Bioinformatics 7
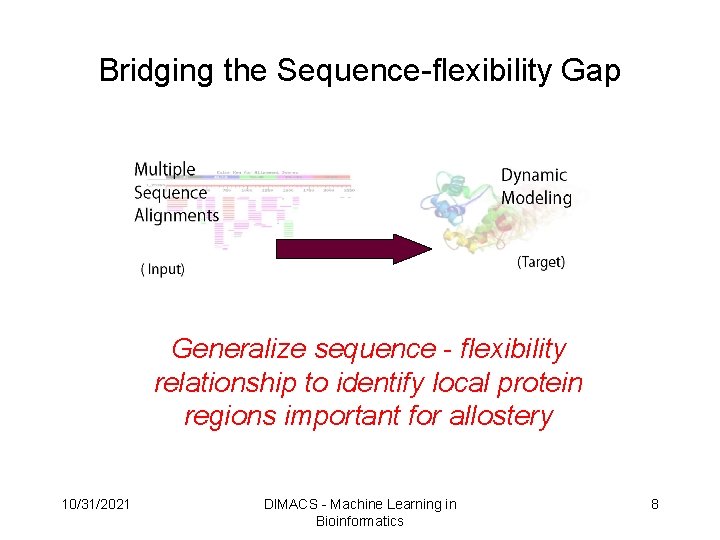
Bridging the Sequence-flexibility Gap Generalize sequence - flexibility relationship to identify local protein regions important for allostery 10/31/2021 DIMACS - Machine Learning in Bioinformatics 8
The Training Dataset The dataset contains the following qualities: • Non-redundant sequences – training set with sequences containing ≤ 10% identity. • With good quality structures – R-factor < 0. 30 • At high resolution – Resolution < 2. 0 Å. Total number of proteins in dataset: 1277 sequences 10/31/2021 DIMACS - Machine Learning in Bioinformatics 9
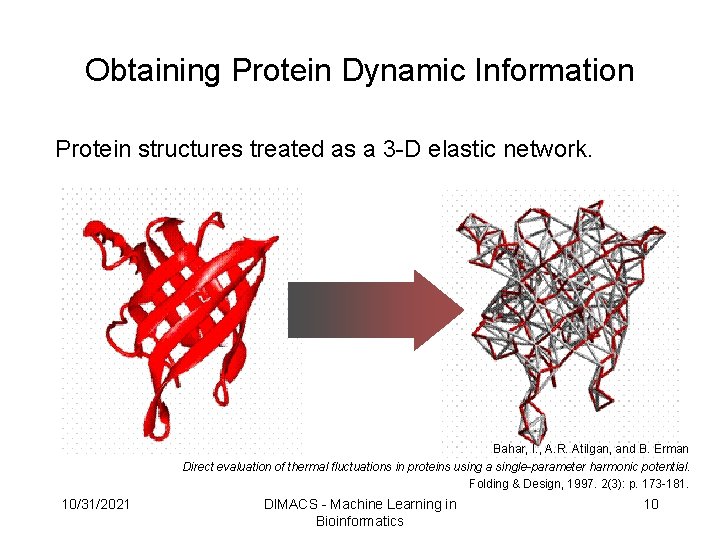
Obtaining Protein Dynamic Information Protein structures treated as a 3 -D elastic network. Bahar, I. , A. R. Atilgan, and B. Erman Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Folding & Design, 1997. 2(3): p. 173 -181. 10/31/2021 DIMACS - Machine Learning in Bioinformatics 10
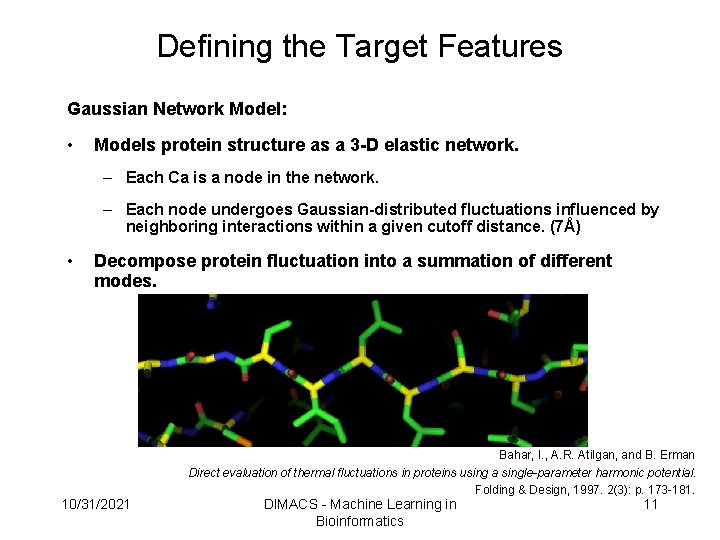
Defining the Target Features Gaussian Network Model: • Models protein structure as a 3 -D elastic network. – Each Ca is a node in the network. – Each node undergoes Gaussian-distributed fluctuations influenced by neighboring interactions within a given cutoff distance. (7Å) • Decompose protein fluctuation into a summation of different modes. Bahar, I. , A. R. Atilgan, and B. Erman Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Folding & Design, 1997. 2(3): p. 173 -181. 10/31/2021 DIMACS - Machine Learning in Bioinformatics 11
Side Note: Gaussian Network Model vs Molecular Dynamics • GNM relatively cause grained • GNM fast to compute vs MD – Look over larger time scales – Suitable for high throughput 10/31/2021 DIMACS - Machine Learning in Bioinformatics 12
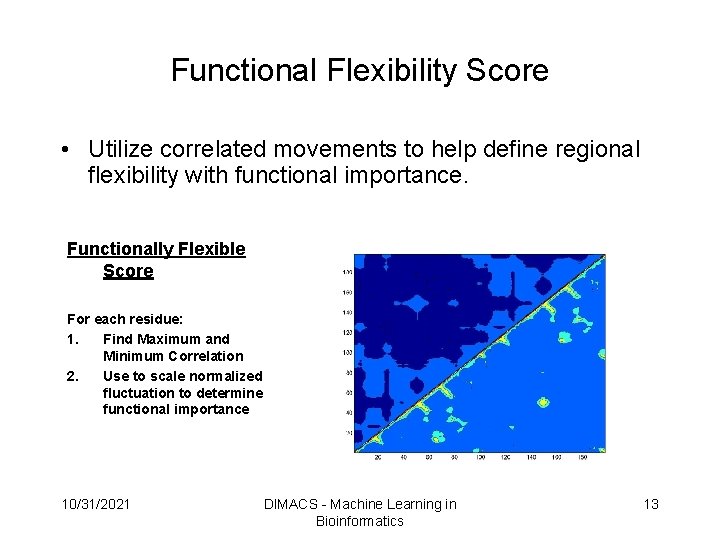
Functional Flexibility Score • Utilize correlated movements to help define regional flexibility with functional importance. Functionally Flexible Score For each residue: 1. Find Maximum and Minimum Correlation 2. Use to scale normalized fluctuation to determine functional importance 10/31/2021 DIMACS - Machine Learning in Bioinformatics 13
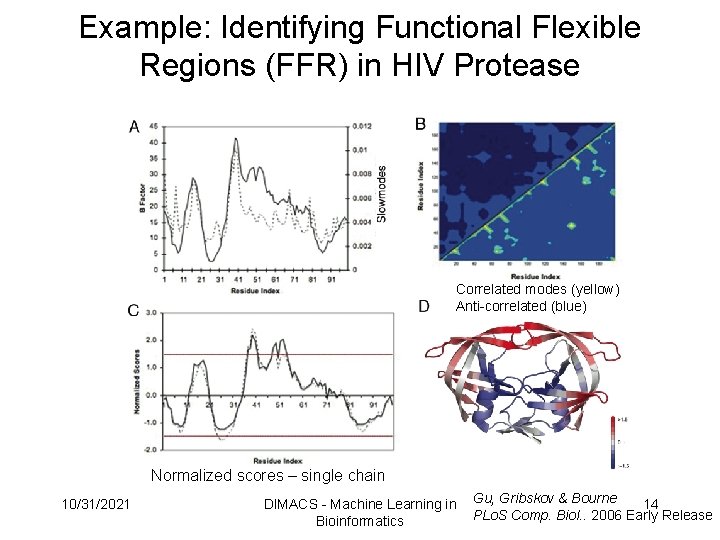
Example: Identifying Functional Flexible Regions (FFR) in HIV Protease Correlated modes (yellow) Anti-correlated (blue) Normalized scores – single chain 10/31/2021 DIMACS - Machine Learning in Bioinformatics Gu, Gribskov & Bourne 14 PLo. S Comp. Biol. . 2006 Early Release
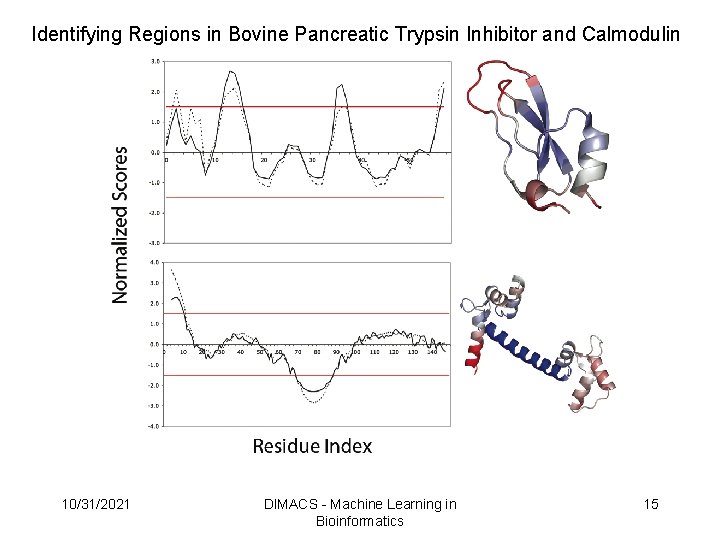
Identifying Regions in Bovine Pancreatic Trypsin Inhibitor and Calmodulin 10/31/2021 DIMACS - Machine Learning in Bioinformatics 15
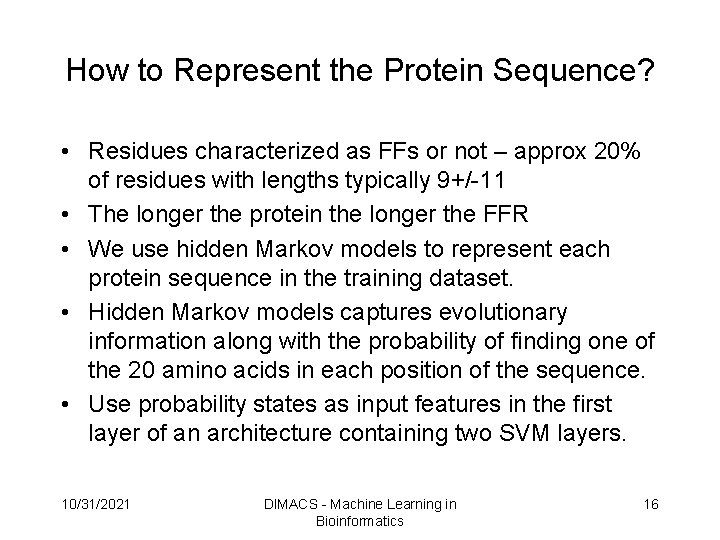
How to Represent the Protein Sequence? • Residues characterized as FFs or not – approx 20% of residues with lengths typically 9+/-11 • The longer the protein the longer the FFR • We use hidden Markov models to represent each protein sequence in the training dataset. • Hidden Markov models captures evolutionary information along with the probability of finding one of the 20 amino acids in each position of the sequence. • Use probability states as input features in the first layer of an architecture containing two SVM layers. 10/31/2021 DIMACS - Machine Learning in Bioinformatics 16
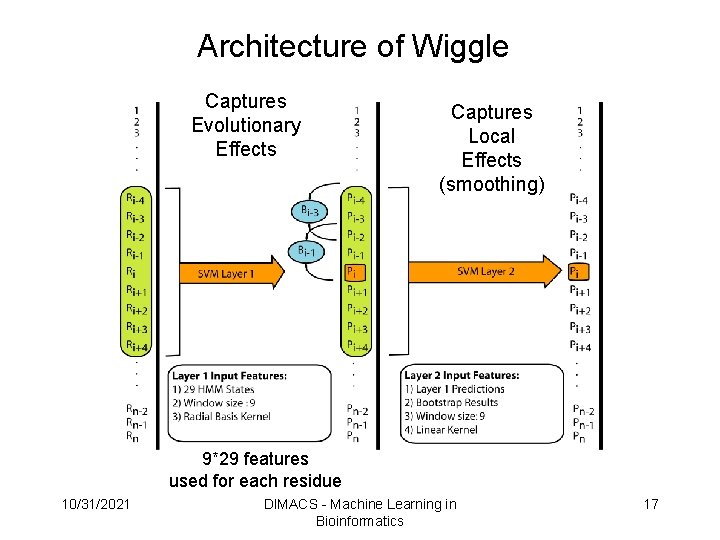
Architecture of Wiggle Captures Evolutionary Effects Captures Local Effects (smoothing) 9*29 features used for each residue 10/31/2021 DIMACS - Machine Learning in Bioinformatics 17
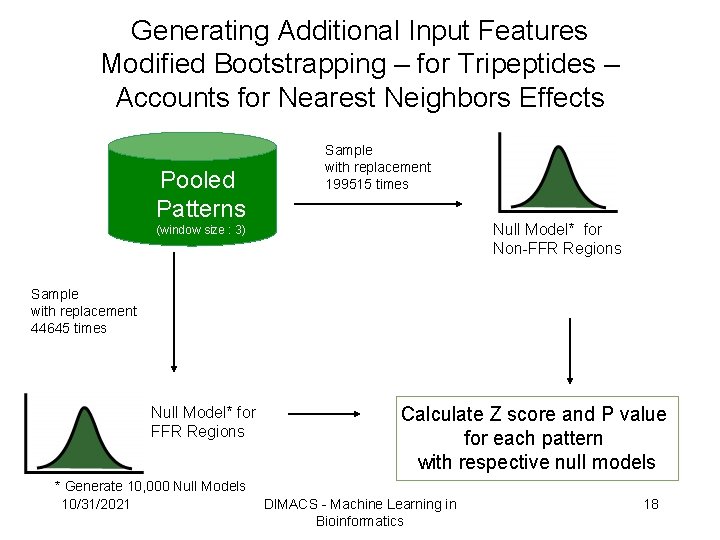
Generating Additional Input Features Modified Bootstrapping – for Tripeptides – Accounts for Nearest Neighbors Effects Pooled Patterns Sample with replacement 199515 times Null Model* for Non-FFR Regions (window size : 3) Sample with replacement 44645 times Null Model* for FFR Regions * Generate 10, 000 Null Models 10/31/2021 Calculate Z score and P value for each pattern with respective null models DIMACS - Machine Learning in Bioinformatics 18
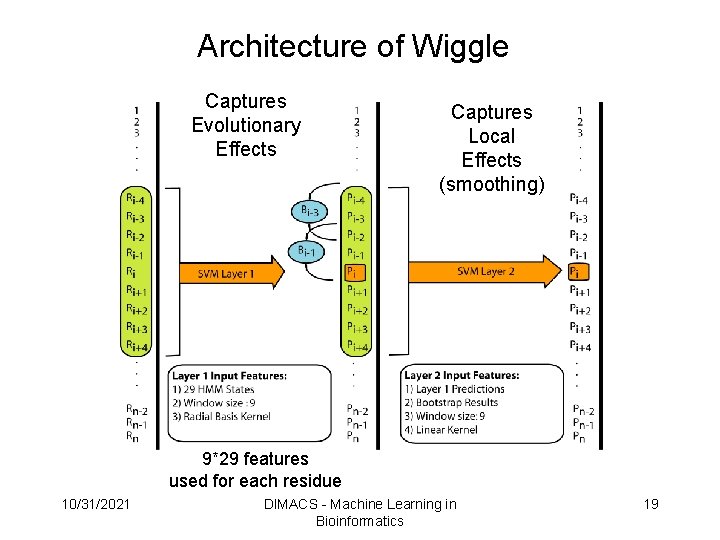
Architecture of Wiggle Captures Evolutionary Effects Captures Local Effects (smoothing) 9*29 features used for each residue 10/31/2021 DIMACS - Machine Learning in Bioinformatics 19
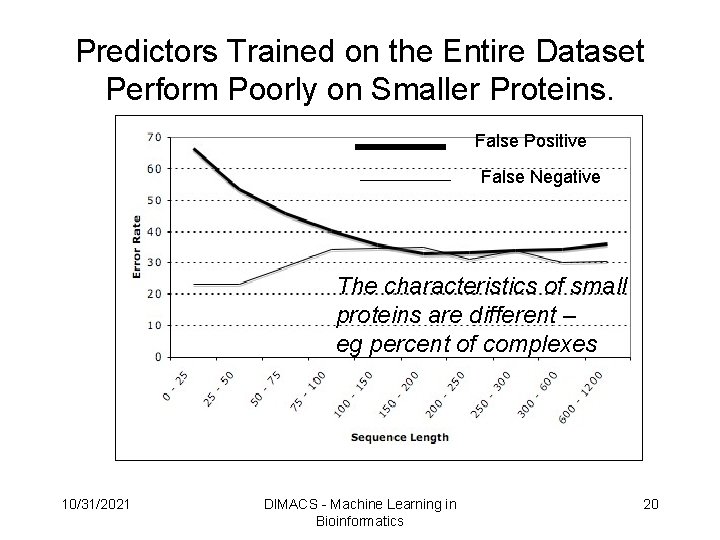
Predictors Trained on the Entire Dataset Perform Poorly on Smaller Proteins. False Positive False Negative The characteristics of small proteins are different – eg percent of complexes 10/31/2021 DIMACS - Machine Learning in Bioinformatics 20
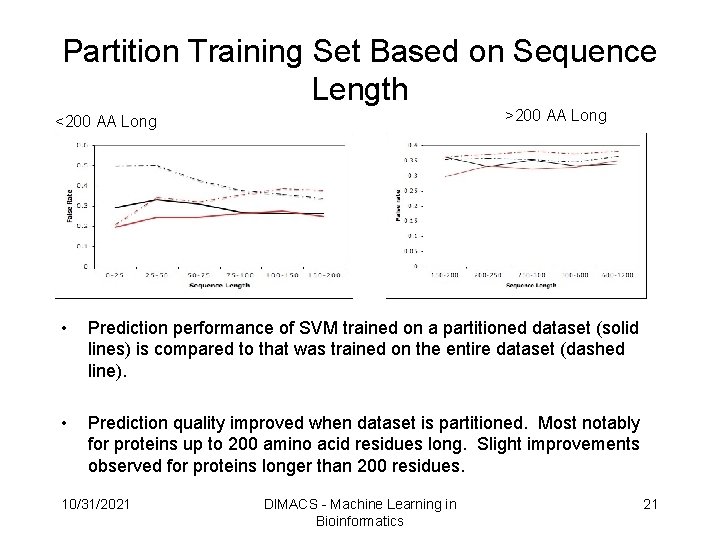
Partition Training Set Based on Sequence Length >200 AA Long <200 AA Long • Prediction performance of SVM trained on a partitioned dataset (solid lines) is compared to that was trained on the entire dataset (dashed line). • Prediction quality improved when dataset is partitioned. Most notably for proteins up to 200 amino acid residues long. Slight improvements observed for proteins longer than 200 residues. 10/31/2021 DIMACS - Machine Learning in Bioinformatics 21
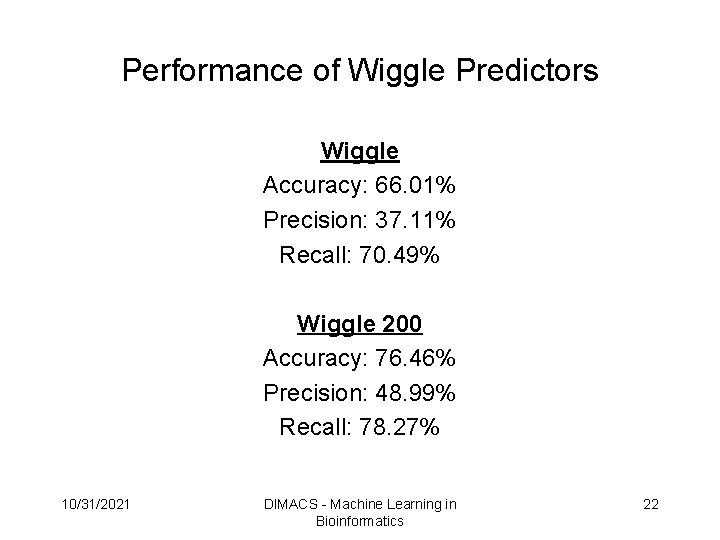
Performance of Wiggle Predictors Wiggle Accuracy: 66. 01% Precision: 37. 11% Recall: 70. 49% Wiggle 200 Accuracy: 76. 46% Precision: 48. 99% Recall: 78. 27% 10/31/2021 DIMACS - Machine Learning in Bioinformatics 22
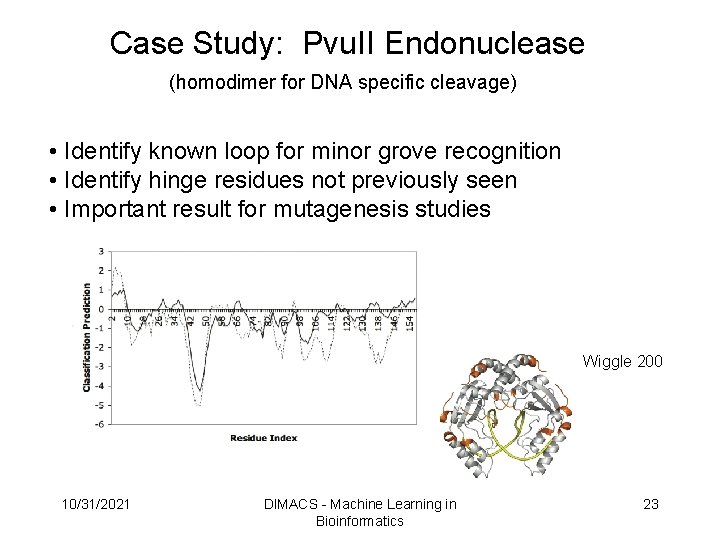
Case Study: Pvu. II Endonuclease (homodimer for DNA specific cleavage) • Identify known loop for minor grove recognition • Identify hinge residues not previously seen • Important result for mutagenesis studies FF SCORE Wiggle 200 10/31/2021 DIMACS - Machine Learning in Bioinformatics 23
Conclusions for Wiggle • FFRs can be measured from structure • With some empirical effort these data can be used as input to an SVM to predict FFRs from sequence alone • Useful for: – – Improving docking studies Better understand protein function Engineer more or less stable proteins …… Gu, Gribskov & Bourne 2006 PLo. S Comp. Biol. . 2006 Early Release 10/31/2021 DIMACS - Machine Learning in Bioinformatics 24
Exploiting Sequence and Structure Homologs to Identify Protein-Protein Binding Sites Jo. Lan Chung, Wang & Bourne 2006 Proteins: Structure, Function and Bioinformatics, 62(3) 630 -640 10/31/2021 DIMACS - Machine Learning in Bioinformatics 25
Methods to Identify Protein-protein Binding Sites • • Docking Threading and homology modeling Evolutionary tracing Correlated mutations Properties of patches Hydrophobicity Neural networks and support vector machines (SVM) 10/31/2021 DIMACS - Machine Learning in Bioinformatics 26

Structurally Conserved Surface Residues? • None of the above methods consider the residues which are spatially conserved on the surfaces of structure homologs • These residues are reported to correspond to the energy hot spots on protein interfaces and can be derived from multiple structure alignments 10/31/2021 DIMACS - Machine Learning in Bioinformatics 27
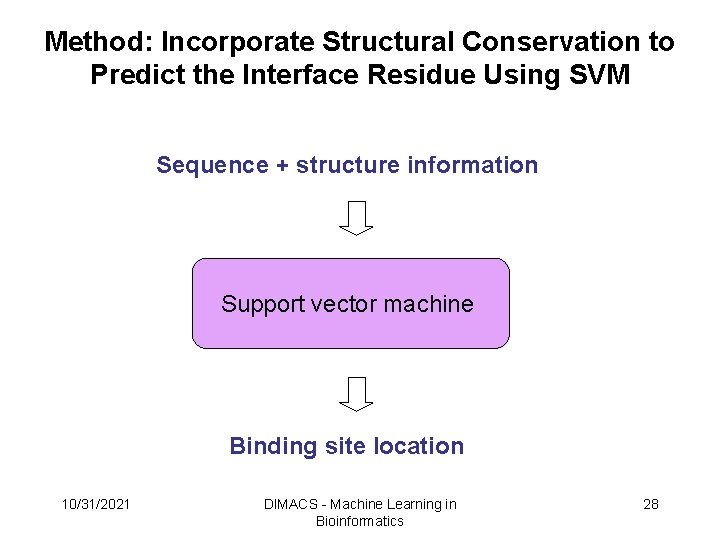
Method: Incorporate Structural Conservation to Predict the Interface Residue Using SVM Sequence + structure information Support vector machine Binding site location 10/31/2021 DIMACS - Machine Learning in Bioinformatics 28
Derive the Structurally Conserved Residues • The structural conservation scores were derived from multiple structural alignments and weighted by the normalized B-factors to consider the structure flexibility that will result in a bad alignment (could use FFRs in the future) • Each position in the alignment has a structural conservation score, which represents the conservation in 3 D space • A position has a high conservation score if the aligned residues are spatially conserved 10/31/2021 DIMACS - Machine Learning in Bioinformatics 29
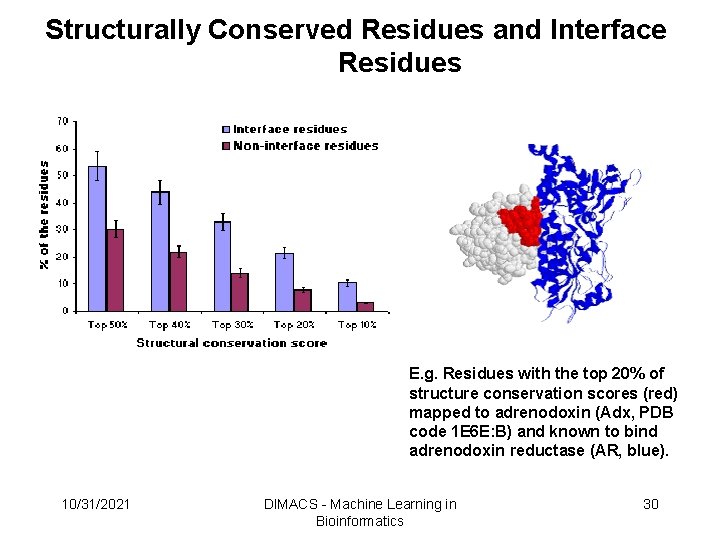
Structurally Conserved Residues and Interface Residues E. g. Residues with the top 20% of structure conservation scores (red) mapped to adrenodoxin (Adx, PDB code 1 E 6 E: B) and known to bind adrenodoxin reductase (AR, blue). 10/31/2021 DIMACS - Machine Learning in Bioinformatics 30
Training Dataset • 274 non-redundant chains of heterocomplexes (<30% sequence identity) extracted from the PDB • Each of these chains was accompanied with a structure alignment with at least 4 members 10/31/2021 DIMACS - Machine Learning in Bioinformatics 31
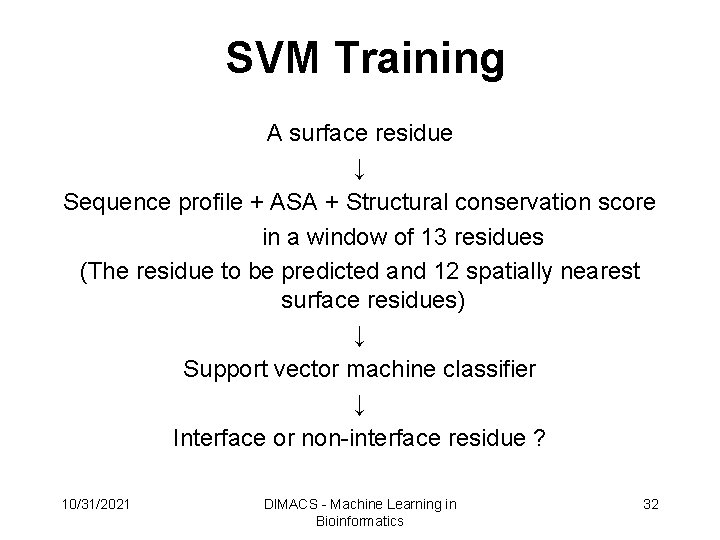
SVM Training A surface residue ↓ Sequence profile + ASA + Structural conservation score in a window of 13 residues (The residue to be predicted and 12 spatially nearest surface residues) ↓ Support vector machine classifier ↓ Interface or non-interface residue ? 10/31/2021 DIMACS - Machine Learning in Bioinformatics 32
SVM Training • Each residue was encoded as a feature vector with 13× 21 dimensions: (the surface residue to be predicted + 12 nearest neighbors) x (20 amino acids + accessible surface area) • Implemented using SVMlight with the radial basis function as a kernel. (γ = 0. 01, regularization parameter C =10) • A set of non-interface surface residues was randomly selected to make the ratio of positive and negative data 1: 1 • 3 fold cross-validation was performed 10/31/2021 DIMACS - Machine Learning in Bioinformatics 33
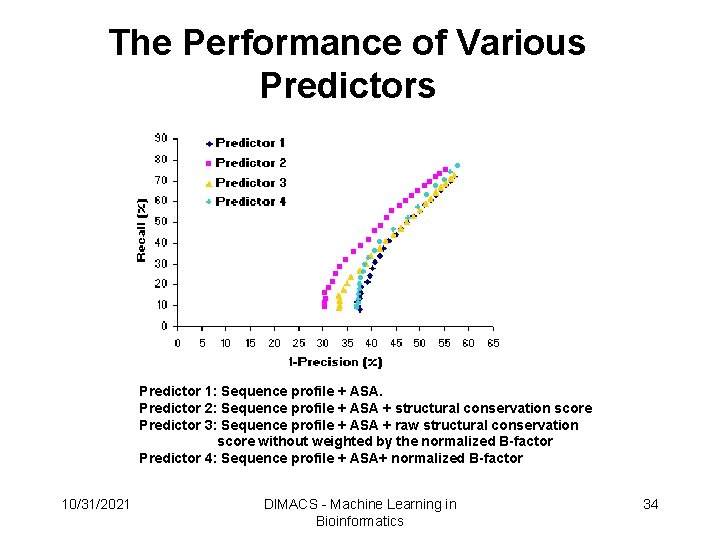
The Performance of Various Predictor 1: Sequence profile + ASA. Predictor 2: Sequence profile + ASA + structural conservation score Predictor 3: Sequence profile + ASA + raw structural conservation score without weighted by the normalized B-factor Predictor 4: Sequence profile + ASA+ normalized B-factor 10/31/2021 DIMACS - Machine Learning in Bioinformatics 34
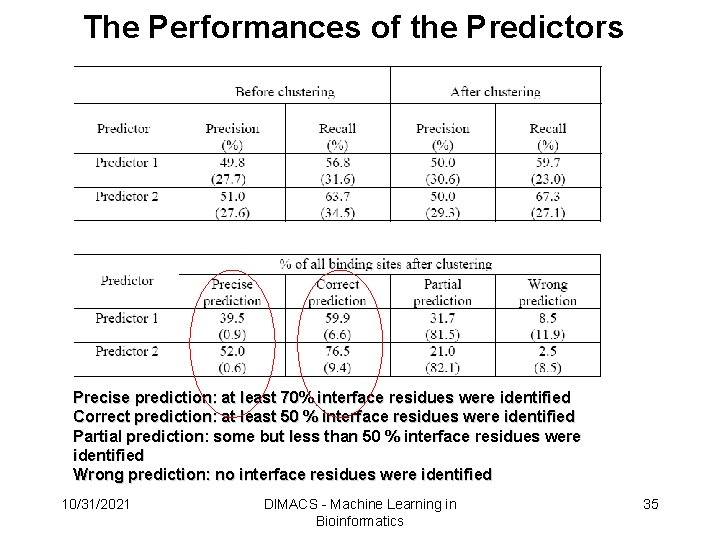
The Performances of the Predictors Precise prediction: at least 70% interface residues were identified Correct prediction: at least 50 % interface residues were identified Partial prediction: some but less than 50 % interface residues were identified Wrong prediction: no interface residues were identified 10/31/2021 DIMACS - Machine Learning in Bioinformatics 35
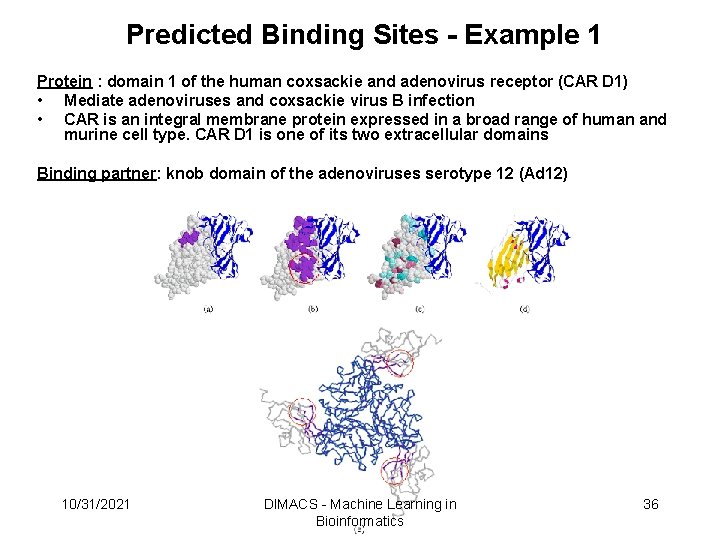
Predicted Binding Sites - Example 1 Protein : domain 1 of the human coxsackie and adenovirus receptor (CAR D 1) • Mediate adenoviruses and coxsackie virus B infection • CAR is an integral membrane protein expressed in a broad range of human and murine cell type. CAR D 1 is one of its two extracellular domains Binding partner: knob domain of the adenoviruses serotype 12 (Ad 12) 10/31/2021 DIMACS - Machine Learning in Bioinformatics 36
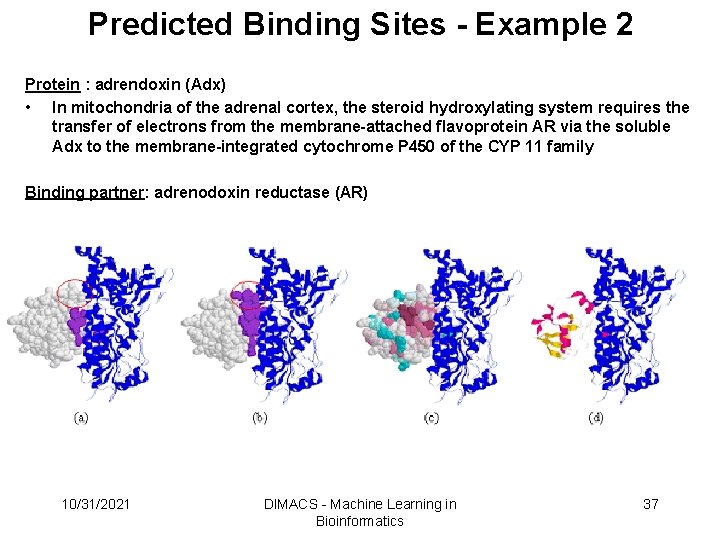
Predicted Binding Sites - Example 2 Protein : adrendoxin (Adx) • In mitochondria of the adrenal cortex, the steroid hydroxylating system requires the transfer of electrons from the membrane-attached flavoprotein AR via the soluble Adx to the membrane-integrated cytochrome P 450 of the CYP 11 family Binding partner: adrenodoxin reductase (AR) 10/31/2021 DIMACS - Machine Learning in Bioinformatics 37
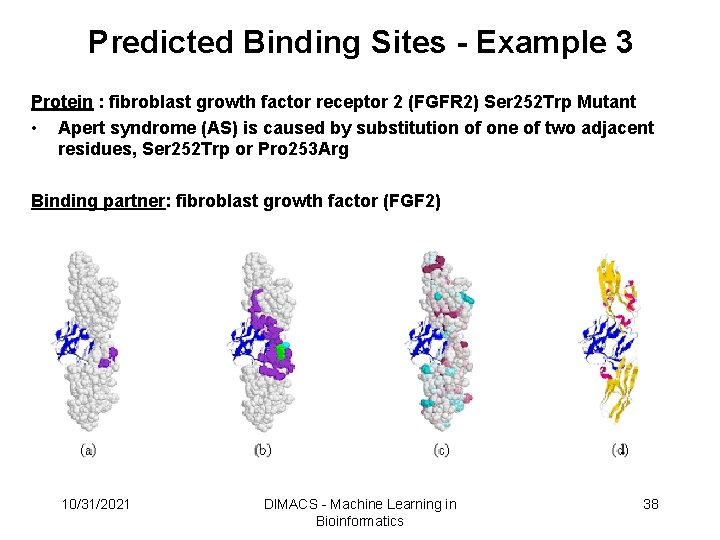
Predicted Binding Sites - Example 3 Protein : fibroblast growth factor receptor 2 (FGFR 2) Ser 252 Trp Mutant • Apert syndrome (AS) is caused by substitution of one of two adjacent residues, Ser 252 Trp or Pro 253 Arg Binding partner: fibroblast growth factor (FGF 2) 10/31/2021 DIMACS - Machine Learning in Bioinformatics 38
Conclusions – Protein-protein Binding Sites • Incorporating the structural conservation score improved the prediction performance of SVM significantly • This study is an initial trial that exploits multiple structure alignment for the large scale prediction of functional regions • We need better algorithms for multiple structure alignment (we have one benchmark for anyone interested) • This method can be used to guide experiments, such as site-specific mutagenesis, or combined with docking procedures to limit the search space 10/31/2021 DIMACS - Machine Learning in Bioinformatics 39
General Conclusions • Using known features of protein structure these can be mapped to the corresponding sequences and used to train an SVM • Having evaluated the SVM in a cross validation tests the performance can be determined • Good performance is shown in training for both flexibility and sites of protein-protein interaction • These predictors are currently being used to solve real biological problems • Can this approach be applied to other aspects of structure? 10/31/2021 DIMACS - Machine Learning in Bioinformatics 40
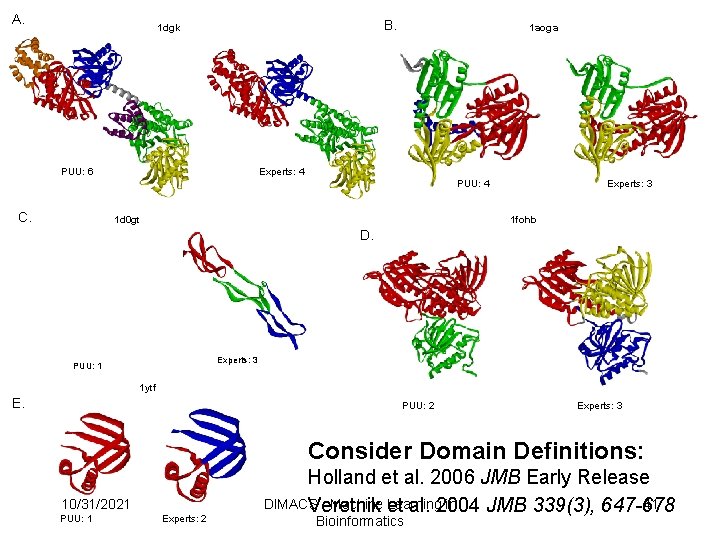
A. B. 1 dgk PUU: 6 1 aoga Experts: 4 PUU: 4 C. 1 d 0 gt Experts: 3 1 fohb D. Experts: 3 PUU: 1 1 ytf E. PUU: 2 Experts: 3 Consider Domain Definitions: 10/31/2021 PUU: 1 Experts: 2 Holland et al. 2006 JMB Early Release DIMACS - Machine et Learning in 41 Veretnik al. 2004 JMB 339(3), 647 -678 Bioinformatics
Challenge – Defining Domain Boundaries from Sequence • A domain is the unit of currency of proteins – domain structures define function, indicate evolutionary relationships etc… • Domain prediction from structure easier than from sequence, but still not a solved problem • Recently developed an accurate test set of domain definitions and boundaries: http: //pdomains. sdsc. edu • Good luck! Benchmark Data Available See: Holland et al 2006 JMB Early Release 10/31/2021 DIMACS - Machine Learning in Bioinformatics 42
Acknowledgements • Functional Flexibility – Jenny Gu & Michael Gribskov • Protein-protein Interactions – Jo. Lan Chung & Wei Wang • Domain Definitions – Stella Veretnik, Tim Holland, Ilya Shindalov, Nick Alexandrov, Abdur Sikur • Funding, NSF, NIH 10/31/2021 DIMACS - Machine Learning in Bioinformatics 43
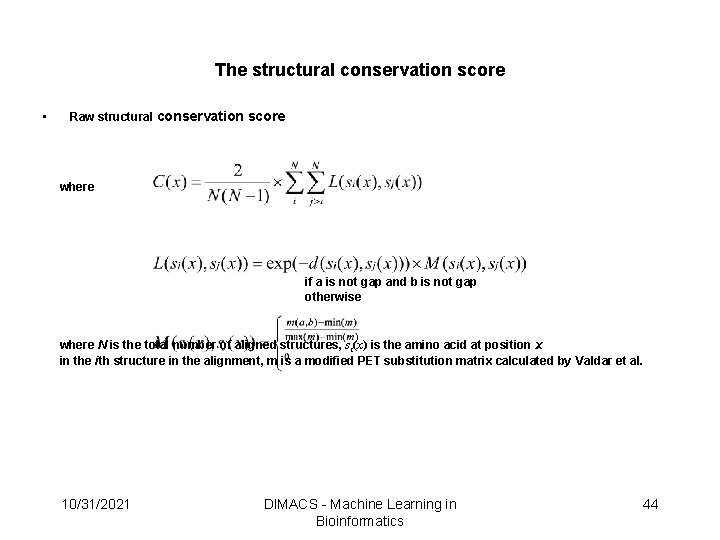
The structural conservation score • Raw structural conservation score where if a is not gap and b is not gap otherwise where N is the total number of aligned structures, si(x) is the amino acid at position x in the ith structure in the alignment, m is a modified PET substitution matrix calculated by Valdar et al. 10/31/2021 DIMACS - Machine Learning in Bioinformatics 44
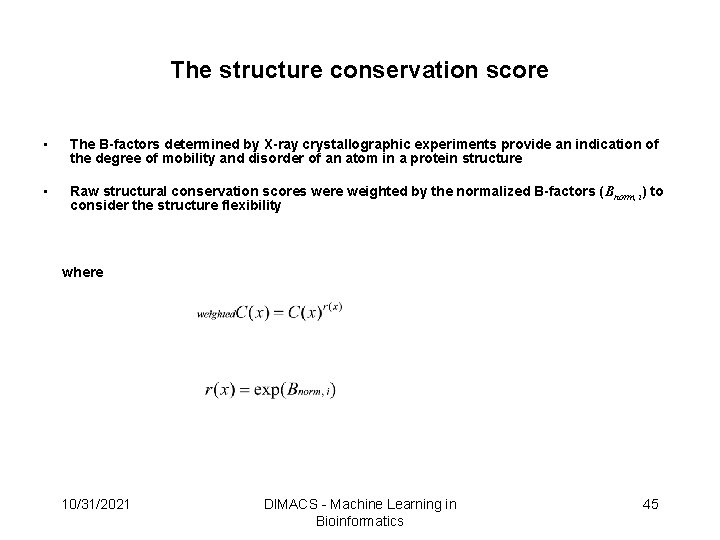
The structure conservation score • The B-factors determined by X-ray crystallographic experiments provide an indication of the degree of mobility and disorder of an atom in a protein structure • Raw structural conservation scores were weighted by the normalized B-factors (Bnorm, i) to consider the structure flexibility where 10/31/2021 DIMACS - Machine Learning in Bioinformatics 45
- Slides: 45