Literature Data Mining and Protein Ontology Development At






- Slides: 18

Literature Data Mining and Protein Ontology Development At the Protein Information Resource (PIR) Hu ZZ*, Mani I, Liu H, Hermoso V, Vijay-Shanker K, Nikolskaya A, Natale DA, and Wu CH ISMB 2005, Detroit, Michigan June 29, 2005 Zhang-Zhi Hu, M. D. Senior Bioinformatics Scientist, PIR Georgetown University Medical Center Washington, DC 20007

PIR – Integrated Protein Informatics Resource for Genomic/Proteomic Research (http: //pir. georgetown. edu) New version of PIR homepage Uni. Prot – Central international database of protein sequence and function (http: //www. uniprot. org) 2

Objective: Accurate, Consistent, and Rich Annotation of Protein Sequence and Function l Literature-Based Curation – Extract Reliable Information from Literature l l Function, domains/sites, developmental stages, catalytic activity, binding and modified residues, regulation, pathways, tissue specificity, subcellular location …. . . Ensure high quality, accurate and up-to-date experimental data for each protein. A major bottleneck! Ontologies/Controlled Vocabularies – For Information Integration and Knowledge Management l Uni. Prot. KB entries will be annotated using widely accepted biological ontologies and other controlled vocabularies, e. g. Gene Ontology (GO) and EC nomenclature. 3

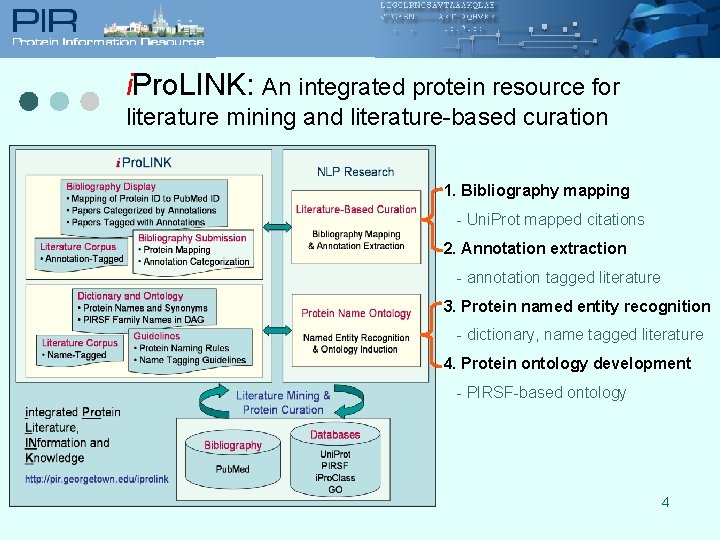
i. Pro. LINK: An integrated protein resource for literature mining and literature-based curation 1. Bibliography mapping - Uni. Prot mapped citations 2. Annotation extraction - annotation tagged literature 3. Protein named entity recognition - dictionary, name tagged literature 4. Protein ontology development - PIRSF-based ontology 4

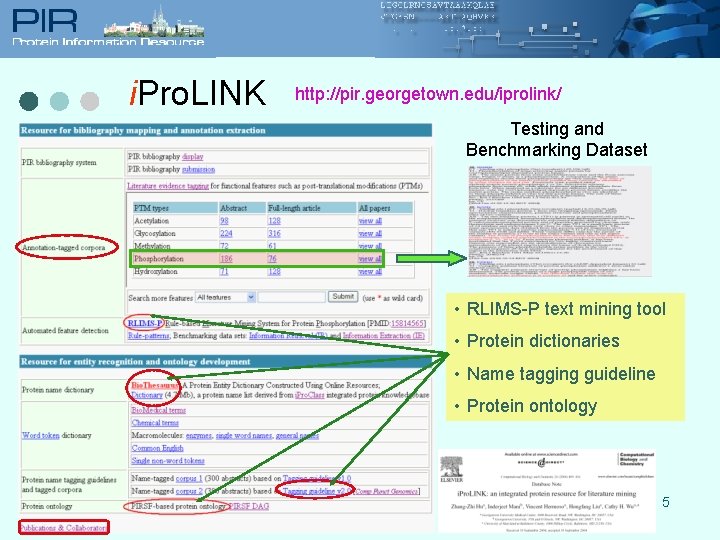
i. Pro. LINK http: //pir. georgetown. edu/iprolink/ Testing and Benchmarking Dataset • RLIMS-P text mining tool • Protein dictionaries • Name tagging guideline • Protein ontology 5

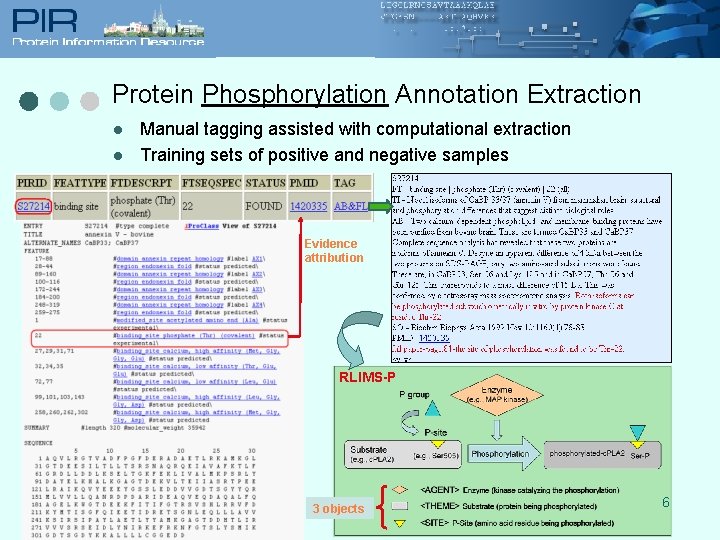
Protein Phosphorylation Annotation Extraction l l Manual tagging assisted with computational extraction Training sets of positive and negative samples Evidence attribution RLIMS-P 3 objects 6

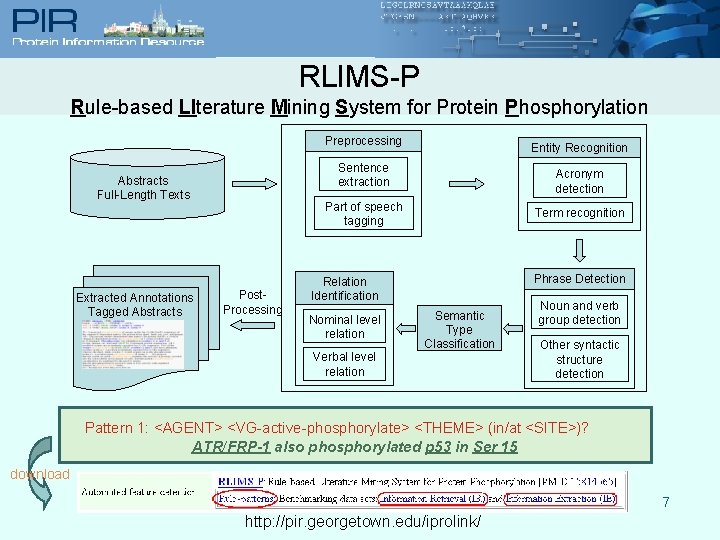
RLIMS-P Rule-based LIterature Mining System for Protein Phosphorylation Preprocessing Sentence extraction Abstracts Full-Length Texts Extracted Annotations Tagged Abstracts Entity Recognition Acronym detection Part of speech tagging Post. Processing Term recognition Phrase Detection Relation Identification Nominal level relation Verbal level relation Semantic Type Classification Noun and verb group detection Other syntactic structure detection Pattern 1: <AGENT> <VG-active-phosphorylate> <THEME> (in/at <SITE>)? ATR/FRP-1 also phosphorylated p 53 in Ser 15 download 7 http: //pir. georgetown. edu/iprolink/

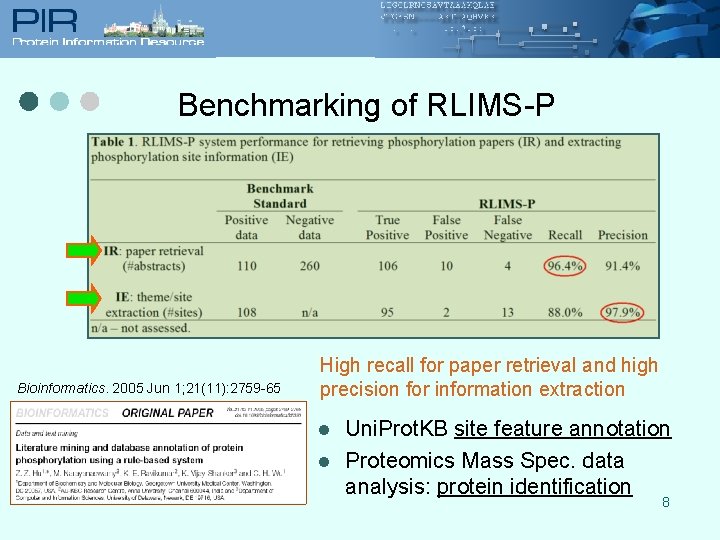
Benchmarking of RLIMS-P Bioinformatics. 2005 Jun 1; 21(11): 2759 -65 High recall for paper retrieval and high precision for information extraction l l Uni. Prot. KB site feature annotation Proteomics Mass Spec. data analysis: protein identification 8

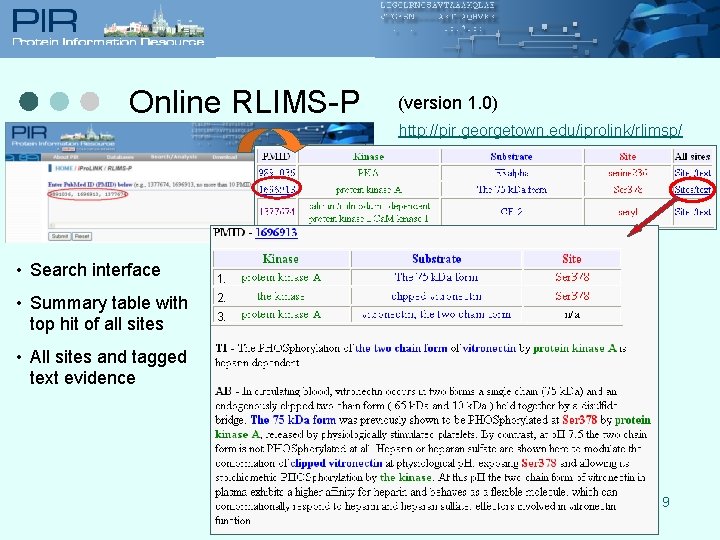
Online RLIMS-P (version 1. 0) http: //pir. georgetown. edu/iprolink/rlimsp/ • Search interface • Summary table with top hit of all sites 1. 2. 3. • All sites and tagged text evidence 9

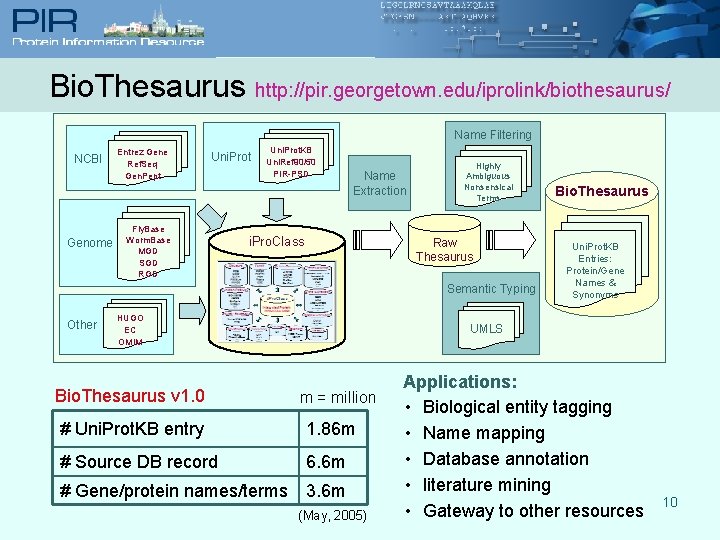
Bio. Thesaurus http: //pir. georgetown. edu/iprolink/biothesaurus/ Name Filtering NCBI Genome Entrez Gene Ref. Seq Gen. Pept Uni. Prot Fly. Base Worm. Base MGD SGD RGD Uni. Prot. KB Uni. Ref 90/50 PIR-PSD Name Extraction i. Pro. Class Highly Ambiguous Nonsensical Terms Raw Thesaurus Semantic Typing Other HUGO EC OMIM Bio. Thesaurus v 1. 0 Bio. Thesaurus Uni. Prot. KB Entries: Protein/Gene Names & Synonyms UMLS m = million # Uni. Prot. KB entry 1. 86 m # Source DB record 6. 6 m # Gene/protein names/terms 3. 6 m (May, 2005) Applications: • Biological entity tagging • Name mapping • Database annotation • literature mining • Gateway to other resources 10

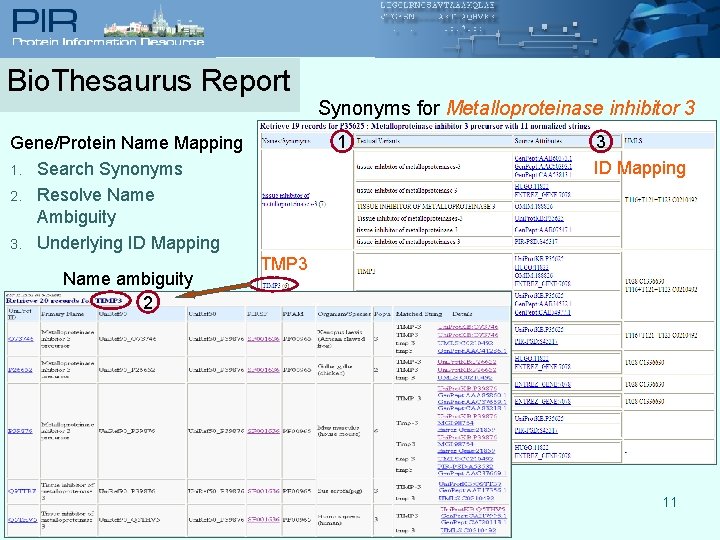
Bio. Thesaurus Report 1 Gene/Protein Name Mapping 1. Search Synonyms 2. Resolve Name Ambiguity 3. Underlying ID Mapping Name ambiguity 2 Synonyms for Metalloproteinase inhibitor 3 3 ID Mapping TMP 3 11

Protein Name Tagging l Tagging guideline versions 1. 0 and 2. 0 l l l Dictionary pre-tagging l l l Generation of domain expert-tagged corpora Inter-coder reliability – upper bound of machine tagging F-measure: 0. 412 (0. 372 Precision, 0. 462 Recall) Advantages: helpful with standardization and extent of tagging, reducing the fatigue problem, and improve intercoder reliability. Bio. Thesaurus for pre-tagging 12

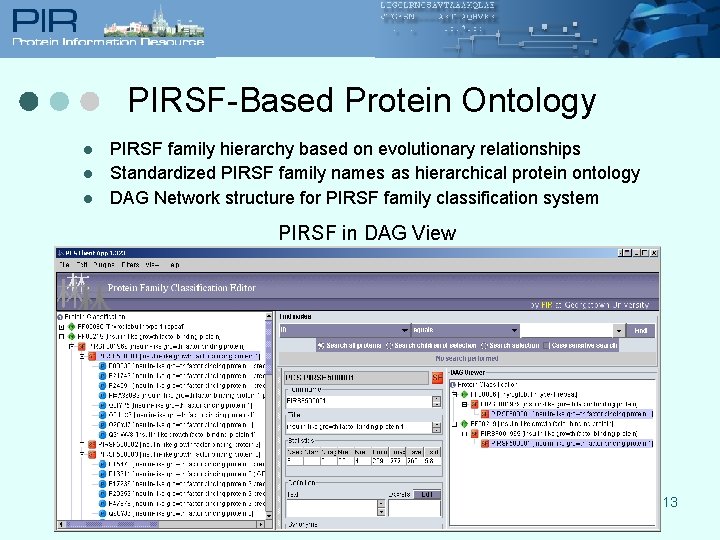
PIRSF-Based Protein Ontology l l l PIRSF family hierarchy based on evolutionary relationships Standardized PIRSF family names as hierarchical protein ontology DAG Network structure for PIRSF family classification system PIRSF in DAG View 13

PIRSF to GO Mapping l Mapped 5363 curated PIRSF homeomorphic families and subfamilies to the GO hierarchy l l l 68% of the PIRSF families and subfamilies map to GO leaf nodes 2329 PIRSFs have shared GO leaf nodes Complements GO: PIRSF-based ontology can be used to analyze GO branches and concepts and to provide links between the GO sub-ontologies Dyn. GO viewer Hongfang Liu University of Maryland l l Superimpose GO and PIRSF hierarchies Bidirectional display (GOor PIRSF-centric views) 14

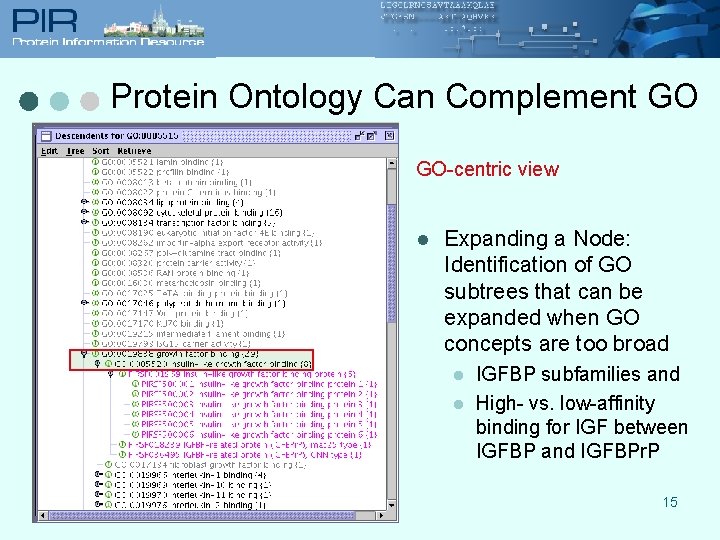
Protein Ontology Can Complement GO GO-centric view l Expanding a Node: Identification of GO subtrees that can be expanded when GO concepts are too broad l l IGFBP subfamilies and High- vs. low-affinity binding for IGF between IGFBP and IGFBPr. P 15

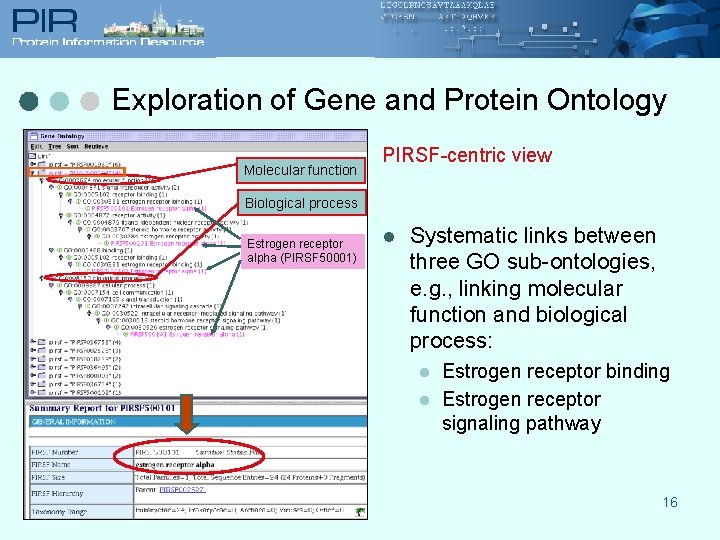
Exploration of Gene and Protein Ontology Molecular function PIRSF-centric view Biological process Estrogen receptor alpha (PIRSF 50001) l Systematic links between three GO sub-ontologies, e. g. , linking molecular function and biological process: l l Estrogen receptor binding Estrogen receptor signaling pathway 16

Summary l l PIR i. Pro. LINK literature mining resource provides annotated data sets for NLP research on annotation extraction and protein ontology development RLIMS-P text-mining tool for protein phosphorylation from Pub. Med literature. Bio. Thesaurus can be used for name mapping to solve name synonym and ambiguity issues. PIRSF-based protein ontology can complement other biological ontologies such as GO. 17

Acknowledgements l Research Projects l l NIH: NHGRI/NIGMS/NLM/NIMH/NCRR/NIDCR (Uni. Prot) NSF: SEIII (Entity Tagging) NSF: ITR (Ontology) Collaborators l l l I. Mani from Georgetown University Department of Linguistics on protein name recognition and protein name ontology. H. Liu from University of Maryland Department of Information System on protein name recognition and text mining. Vijay K. Shanker from University of Delaware Department of Computer and Information Science on text mining of protein phosphorylation features. 18