Linking Open Drug Data Susie Stephens Principal Research







- Slides: 11

Linking Open Drug Data Susie Stephens, Principal Research Scientist, Eli Lilly

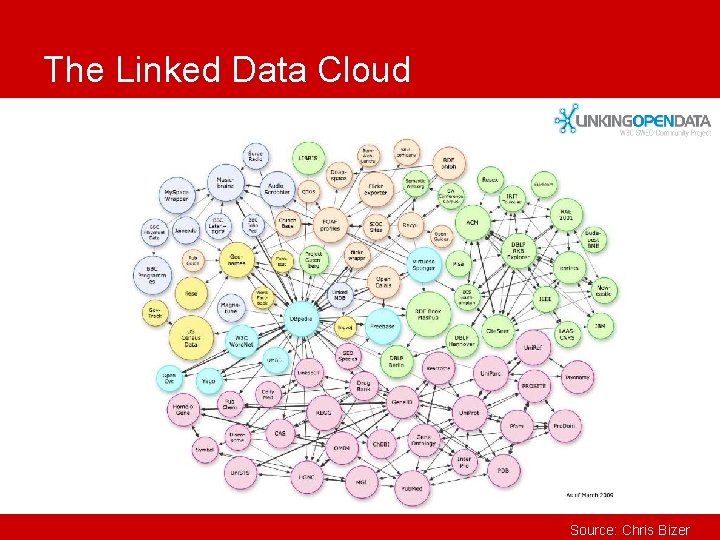
The Linked Data Cloud Source: Chris Bizer

Linking Open Drug Data • HCLSIG task started October 1, 2008 • Primary Objectives • Survey publicly available data sets about drugs • Publish and interlink these data sets on the Web • Explore interesting questions in competitive intelligence that could be answered if the data sets are linked • Participants: Bosse Andersson, Chris Bizer, Kei Cheung, Don Doherty, Oktie Hassanzadeh, Anja Jentzsch, Scott Marshall, Eric Prud’hommeaux, Matthias Samwald, Susie Stephens, Jun Zhao

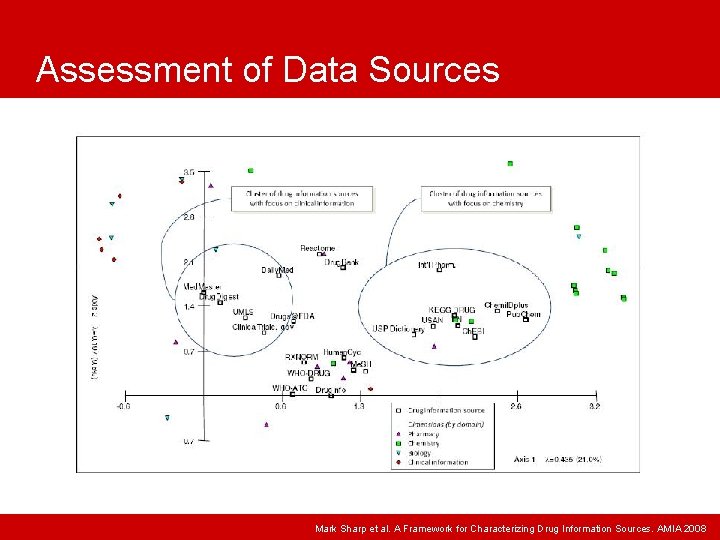
Assessment of Data Sources Mark Sharp et al. A Framework for Characterizing Drug Information Sources. AMIA 2008

Published Data Sets • Linked. CT (http: //linkedct. org) • • • Online registry of more than 60, 000 clinical trials Published in XML 7, 011, 000 triples (290, 000 interlinking) • Drug. Bank (http: //www 4. wiwiss. fu-berlin. de/drugbank) • • • A repository of almost 5, 000 FDA-approved drugs Published as Drug. Bank Drug. Cards 1, 153, 000 triples (23, 000 interlinking) • Daily. Med (http: //www 4. wiwiss. fu-berlin. de/dailymed/) • • • High quality information about marketed drugs Flat file representation 124, 000 triples (29, 600 interlinking) • Diseasome (http: //www 4. wiwiss. fu-berlin. de/diseasome) Information about 4, 300 disorders and disease genes linked by known disorder-gene associations • Published in XML • 88, 000 triples (23, 000 interlinking) •

Classes of Links • Based on common identifiers • Links present in the source data sets • Based on link discovery and record linkage techniques • String matching – E. g. , “Alzheimer’s disease” in Linked. CT was matched with “Alzheimer_disease” in Diseasome • Semantic matching – E. g. “Varenicline” has the synonym “Varenicline Tartrate” and the brand names “Champix” and “Chantix”

Business Use Case • A neuroscience focused business manager is interested in seeing an update on new clinical trials by competitors on Alzheimer’s Disease (AD) • A phase III trial by Pfizer for a drug called Varenicline has just been listed in linked. CT • More information of interest is found in DBpedia, Daily. Med, and Drug. Bank • Daily. Med indicates the drug is already on the market for Nicotine addiction and has minimal side effects • Drug. Bank allows the manager to see the targets for Varenicline • Diseasome, however, indicates that the corresponding genes are only implicated in nicotine addiction, rather than AD • This suggests a more complex relationship between the diseases than just the drug target • Extending the browsing to the SWAN Knowledgebase shows that there are hypotheses relating AD to nicotine receptors through amyloid beta

Technical Challenges • Life sciences data is difficult to connect due to inconsistent terminology and the prevalence of synonyms, and homonyms • Refinement of tools and techniques for enabling more automatic linking of entities across data sets • Selection of ontologies to enable consistent mappings • Development a sufficiently robust platform as to enable inferencing • Provide an interface to users that supports browsing, querying, and filtering data • Persuade data providers to publish in RDF would alleviate the need for us to update data, and provide some of the interlinking

Next Steps • Ensure that existing data are accurately and comprehensively linked • Incorporate additional data sources into the LODD cloud that are of interest to competitive intelligence (e. g. Traditional Chinese Medicine) • Use novel link discovery tools and frameworks including Silk and Lin. Quer • Explore using SIOC to aggregate information as what patients are saying about drugs • Submit paper to the i. Triplify Challenge

Task Alignment • LODD is looking to use Pharma Ontology’s work to help inform the mappings • Data converted to RDF is also loaded into Bio. RDF’s HCLS KB

Conclusions • Added 4 drug-related data sets into the cloud for competitive intelligence • Will add further data sources to the LODD cloud to enable more insights to be gleaned • Will continue to explore and test tools that are being developed for LOD