Linkage The Use of Linkage Learning in Genetic
- Slides: 1

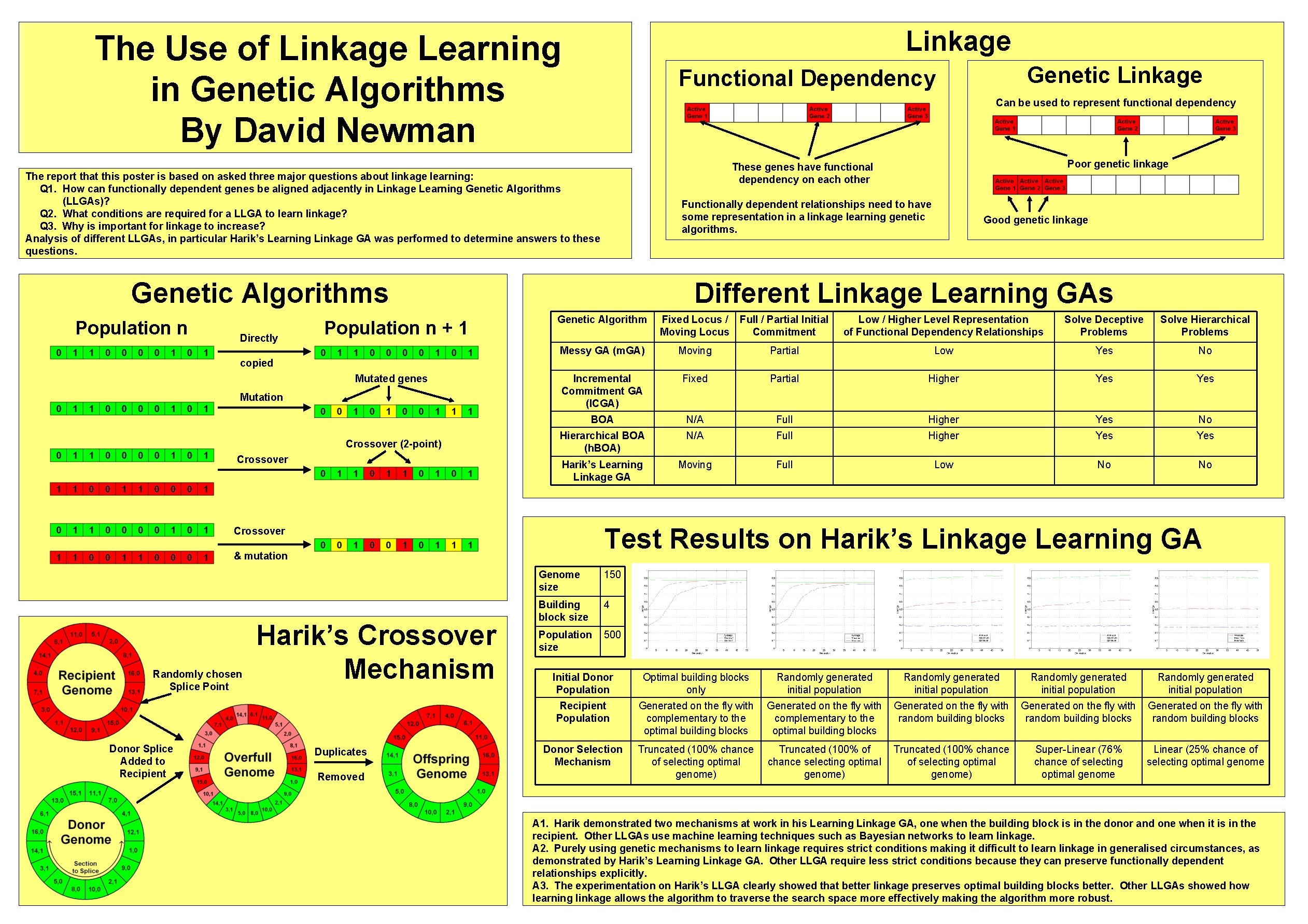
Linkage The Use of Linkage Learning in Genetic Algorithms By David Newman Can be used to represent functional dependency Functionally dependent relationships need to have some representation in a linkage learning genetic algorithms. Genetic Algorithms Directly Population n + 1 Poor genetic linkage These genes have functional dependency on each other The report that this poster is based on asked three major questions about linkage learning: Q 1. How can functionally dependent genes be aligned adjacently in Linkage Learning Genetic Algorithms (LLGAs)? Q 2. What conditions are required for a LLGA to learn linkage? Q 3. Why is important for linkage to increase? Analysis of different LLGAs, in particular Harik’s Learning Linkage GA was performed to determine answers to these questions. Population n Genetic Linkage Functional Dependency Good genetic linkage Different Linkage Learning GAs Genetic Algorithm Fixed Locus / Full / Partial Initial Moving Locus Commitment Low / Higher Level Representation of Functional Dependency Relationships Solve Deceptive Problems Solve Hierarchical Problems Messy GA (m. GA) Moving Partial Low Yes No Incremental Commitment GA (ICGA) Fixed Partial Higher Yes BOA N/A Full Higher Yes No Hierarchical BOA (h. BOA) N/A Full Higher Yes Harik’s Learning Linkage GA Moving Full Low No No copied Mutated genes Mutation Crossover (2 -point) Crossover Test Results on Harik’s Linkage Learning GA Crossover & mutation Randomly chosen Splice Point Donor Splice Added to Recipient Harik’s Crossover Mechanism Duplicates Removed Genome size 150 Building block size 4 Population size 500 Initial Donor Population Optimal building blocks only Randomly generated initial population Recipient Population Generated on the fly with complementary to the optimal building blocks Generated on the fly with random building blocks Donor Selection Mechanism Truncated (100% chance of selecting optimal genome) Truncated (100% of chance selecting optimal genome) Truncated (100% chance of selecting optimal genome) Super-Linear (76% chance of selecting optimal genome Linear (25% chance of selecting optimal genome A 1. Harik demonstrated two mechanisms at work in his Learning Linkage GA, one when the building block is in the donor and one when it is in the recipient. Other LLGAs use machine learning techniques such as Bayesian networks to learn linkage. A 2. Purely using genetic mechanisms to learn linkage requires strict conditions making it difficult to learn linkage in generalised circumstances, as demonstrated by Harik’s Learning Linkage GA. Other LLGA require less strict conditions because they can preserve functionally dependent relationships explicitly. A 3. The experimentation on Harik’s LLGA clearly showed that better linkage preserves optimal building blocks better. Other LLGAs showed how learning linkage allows the algorithm to traverse the search space more effectively making the algorithm more robust.