LINEs SINEs 1 The genome of most eukaryotes




















- Slides: 67

LINEs & SINEs 1

The genome of most eukaryotes contains highly-repetitive interspersed sequences: sequences (1) short-interspersed repetitive elements (SINEs) SINE (2) long-interspersed repetitive elements (LINEs). LINE 2

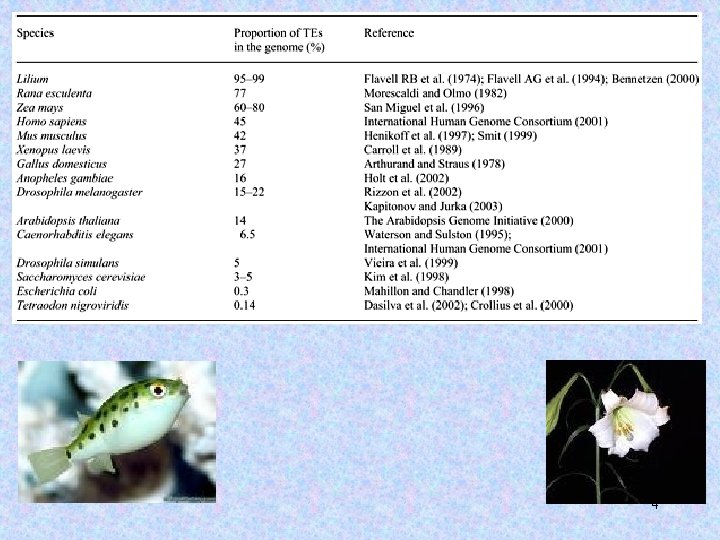
~1/2 of the human genome consists of interspersed repetitive sequences. 3

4

5

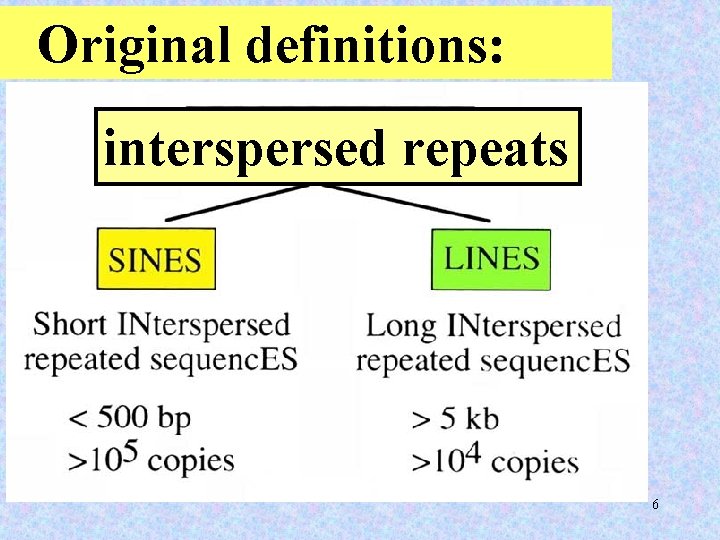
Original definitions: interspersed repeats 6

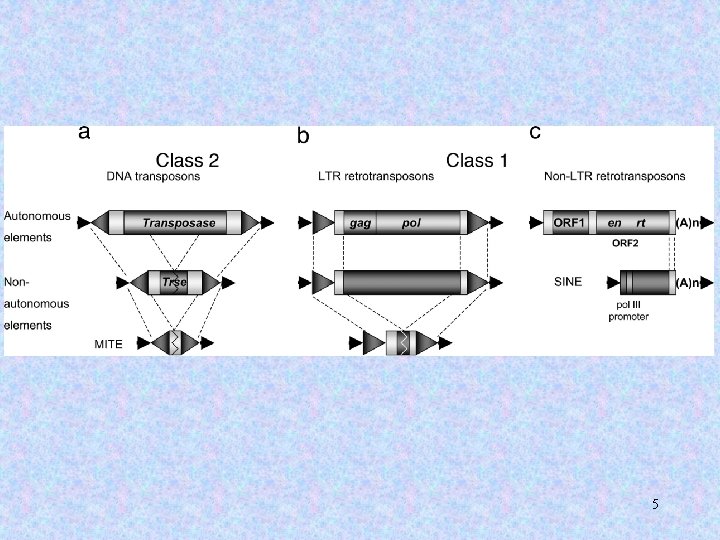
Current definitions: LINEs = Active or degenerate descendants of transposable elements. SINEs = Non-autonomous transposable elements (lacking the ability to mediate their own transposition) and their degenerate descendents. 7

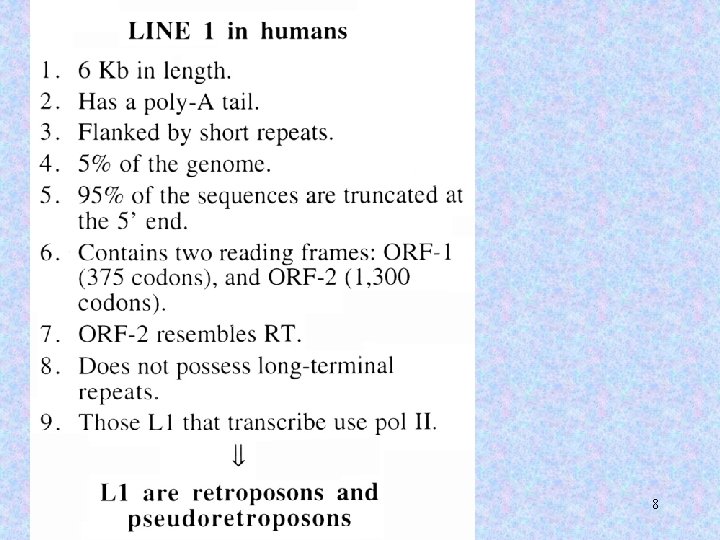
8

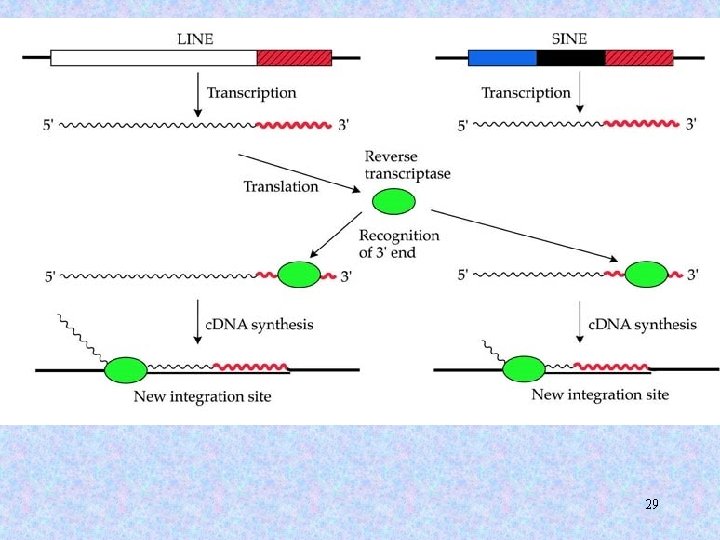
The reverse transcriptase has LINE specificity, i. e. , a reverse transcriptase from one LINE will only recognize the 3’ end of that LINE, and will be less efficient at recognizing and reverse transcribing other LINEs. 9

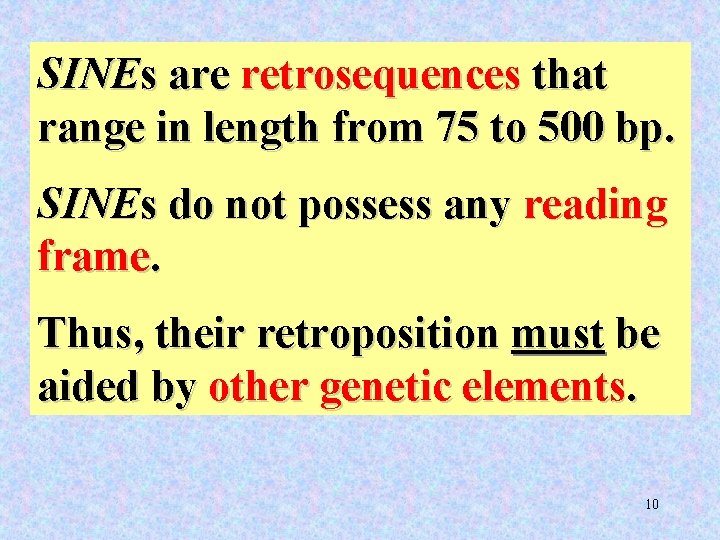
SINEs are retrosequences that range in length from 75 to 500 bp. SINEs do not possess any reading frame. Thus, their retroposition must be aided by other genetic elements. 10

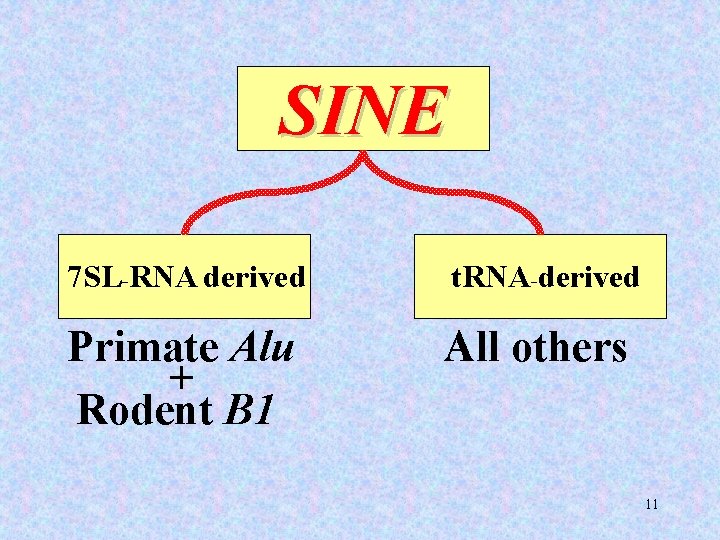
SINE 7 SL-RNA derived t. RNA-derived Primate Alu + Rodent B 1 All others 11

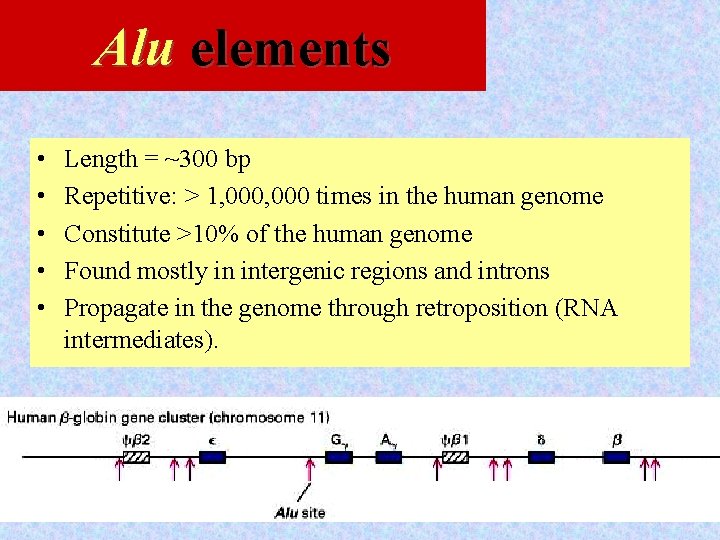
Alu elements • • • Length = ~300 bp Repetitive: > 1, 000 times in the human genome Constitute >10% of the human genome Found mostly in intergenic regions and introns Propagate in the genome through retroposition (RNA intermediates). 12

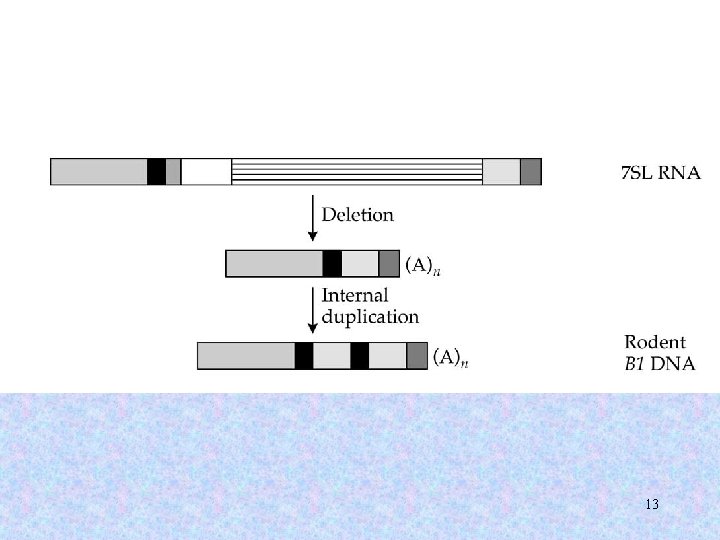
13

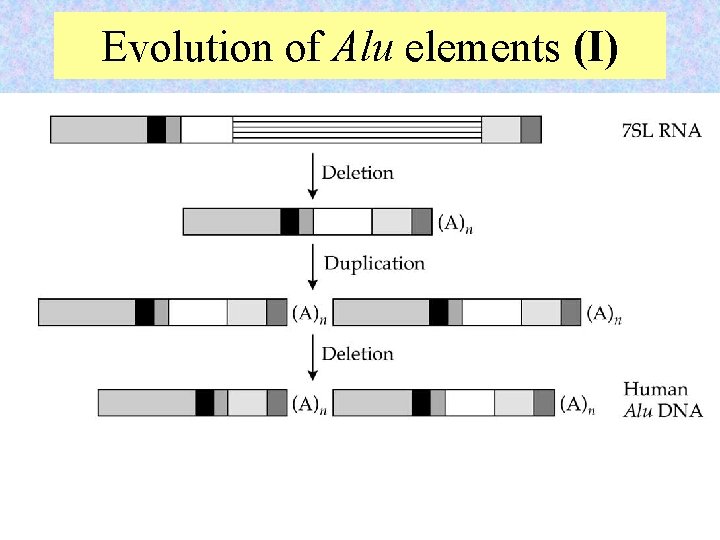
Evolution of Alu elements (I) 14

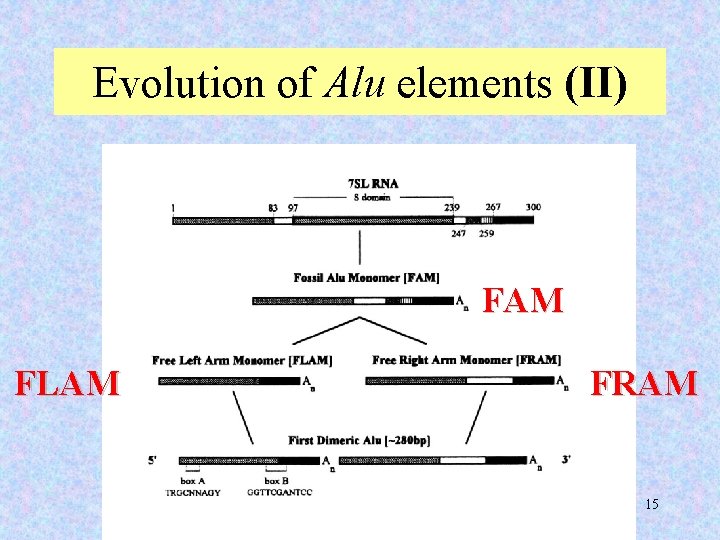
Evolution of Alu elements (II) FAM FLAM FRAM 15

16

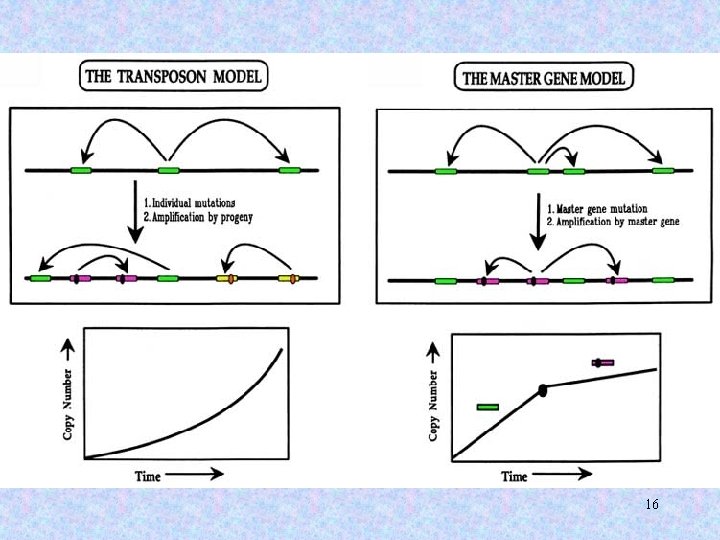
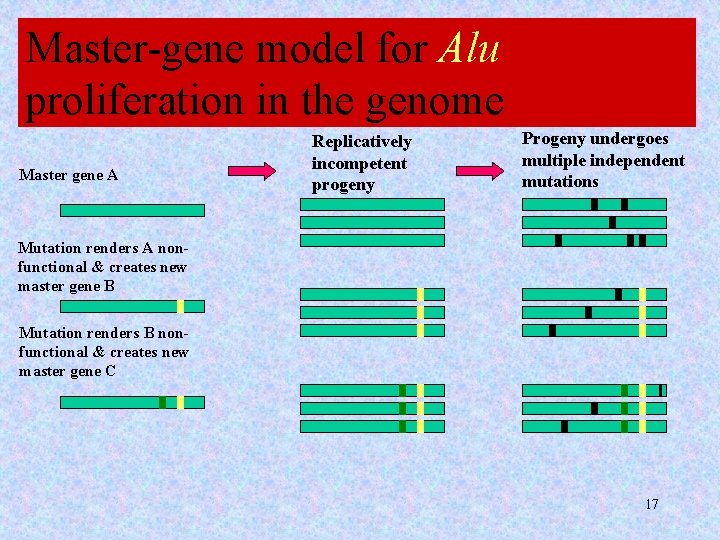
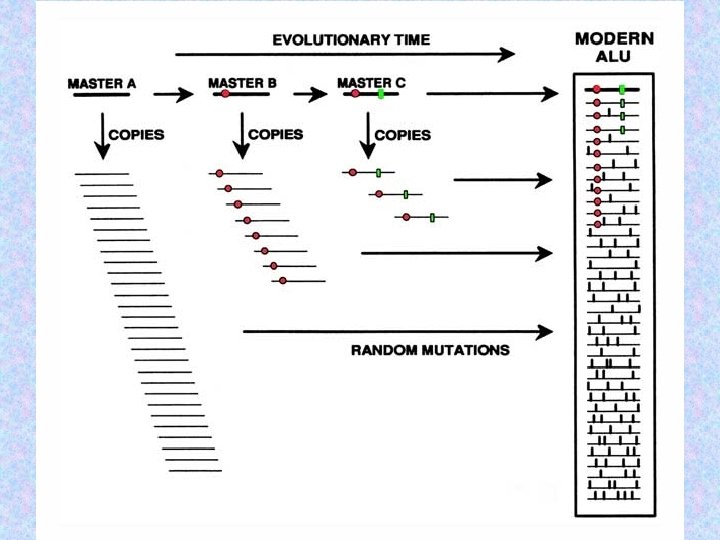
Master-gene model for Alu proliferation in the genome Master gene A Replicatively incompetent progeny Progeny undergoes multiple independent mutations Mutation renders A nonfunctional & creates new master gene B Mutation renders B nonfunctional & creates new master gene C 17

18

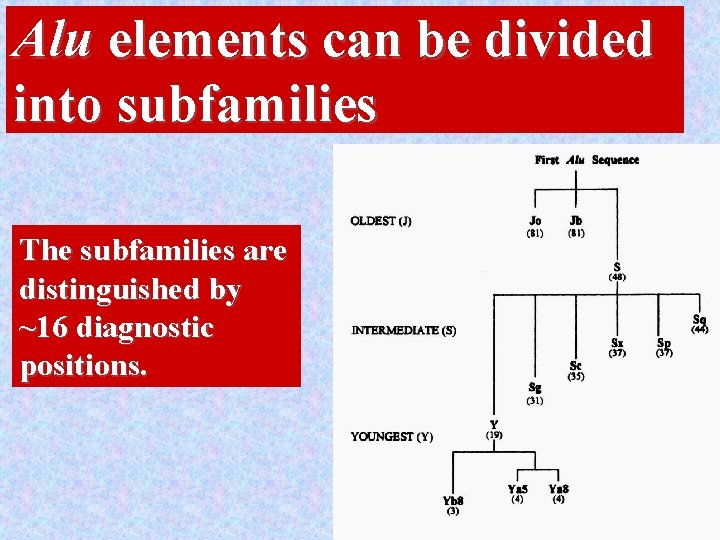
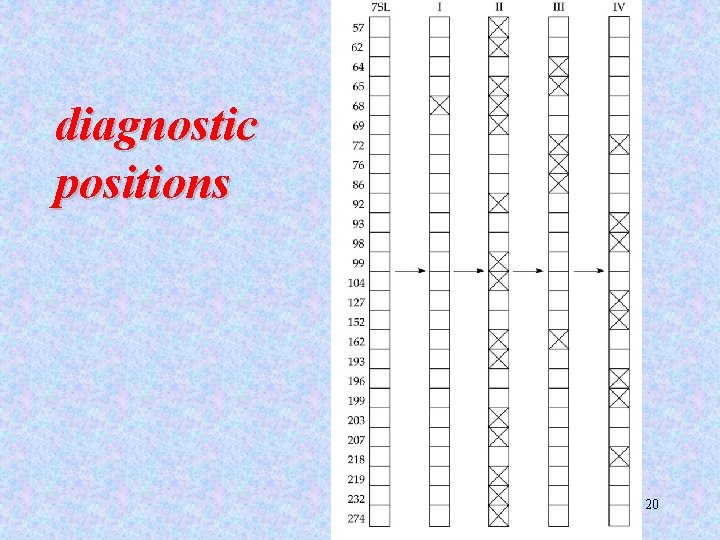
Alu elements can be divided into subfamilies The subfamilies are distinguished by ~16 diagnostic positions. 19

diagnostic positions 20

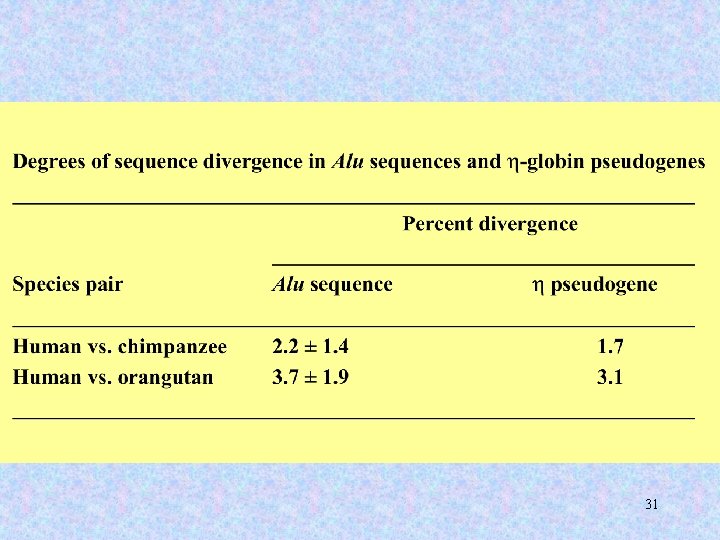
Alu elements are found only in primates. All the millions of Alu elements have accumulated in a mere ~65 million years. 21

Alu elements can be sorted into distinct families according to shared patterns of variation. At any given point in time, only one or several Alu “master copies” are capable of transposing. 22

Early in primate evolution, Alu transposition rate was approximately one new jump in every live birth. Today, it is about one new jump in every 200 live births. 23

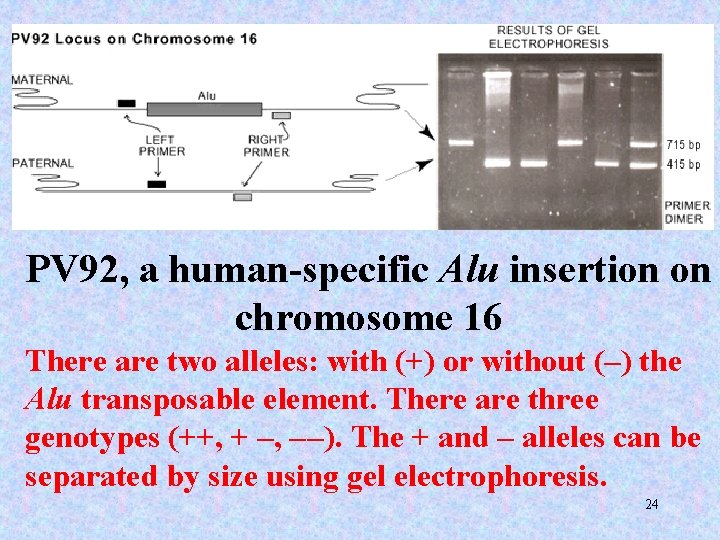
PV 92, a human-specific Alu insertion on chromosome 16 There are two alleles: with (+) or without (–) the Alu transposable element. There are three genotypes (++, + –, – –). The + and – alleles can be separated by size using gel electrophoresis. 24

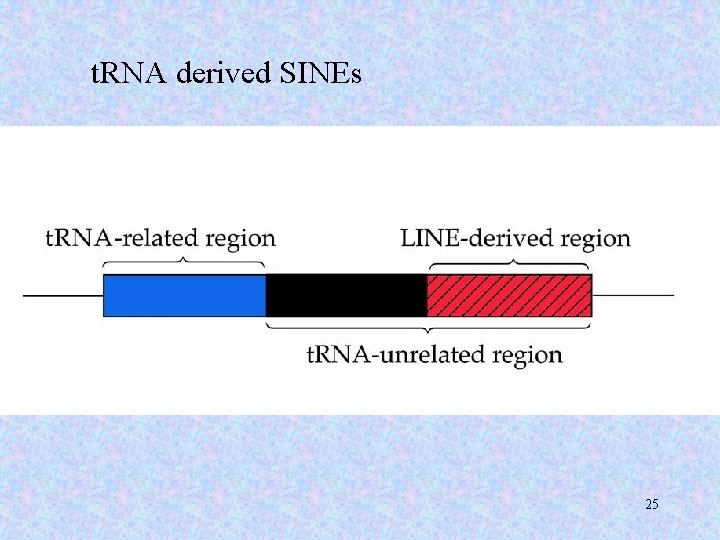
t. RNA derived SINEs 25

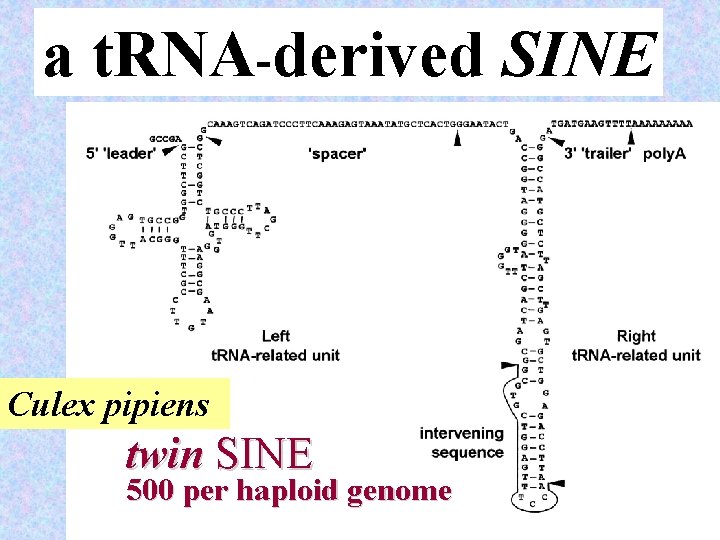
a t. RNA-derived SINE Culex pipiens twin SINE 500 per haploid genome 26

Norihiro Okada et al. 27

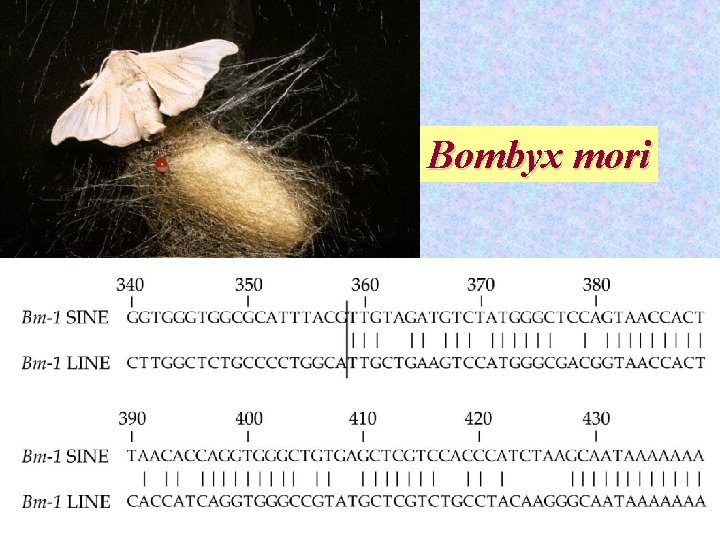
Bombyx mori 28

29

3 predictions: 1. Where there’s a SINE, there’s a LINE! 2. Once a LINE partner becomes inactive, the SINE partner will lose its ability to retrotranspose. 3. A LINE partner should have a longer evolutionary history and, hence, a broader phylogenetic distribution than its SINE partner. 30

31

GENETIC AND EVOLUTIONARY EFFECTS OF TRANSPOSITION 1. Duplicative transposition increases genome size. Lily Edible frog Sunflower 32

2. Bacterial transposons often carry genes that confer antibiotic or other forms of resistance. Plasmids can carry such transposons from cell to cell, so that resistance can spread throughout a population or an ecosystem. 33

34

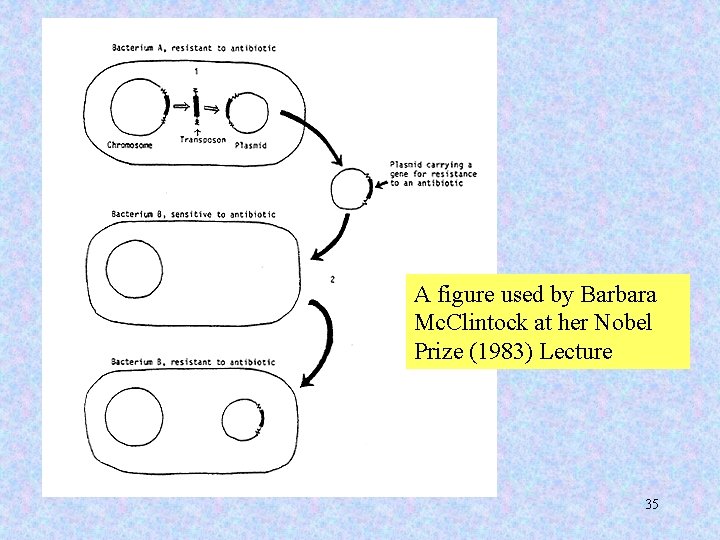
A figure used by Barbara Mc. Clintock at her Nobel Prize (1983) Lecture 35

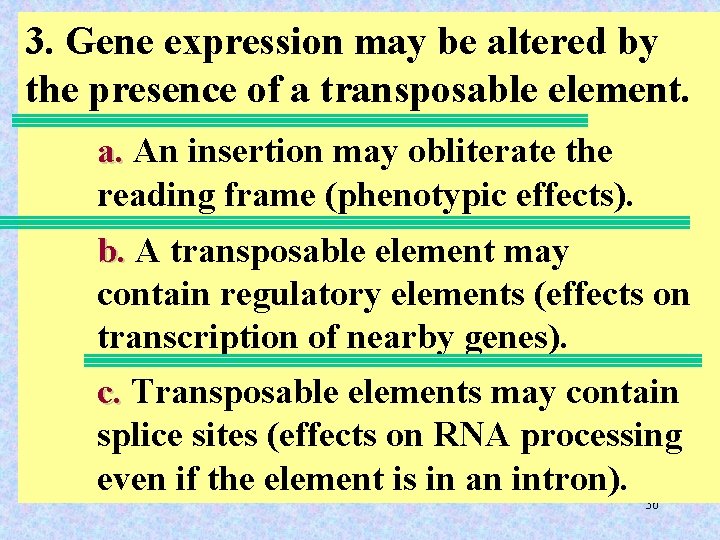
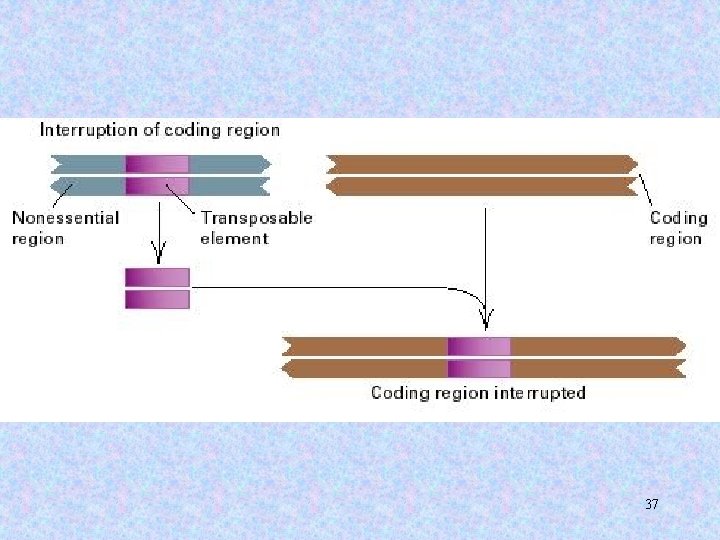
3. Gene expression may be altered by the presence of a transposable element. a. An insertion may obliterate the reading frame (phenotypic effects). b. A transposable element may contain regulatory elements (effects on transcription of nearby genes). c. Transposable elements may contain splice sites (effects on RNA processing even if the element is in an intron). 36

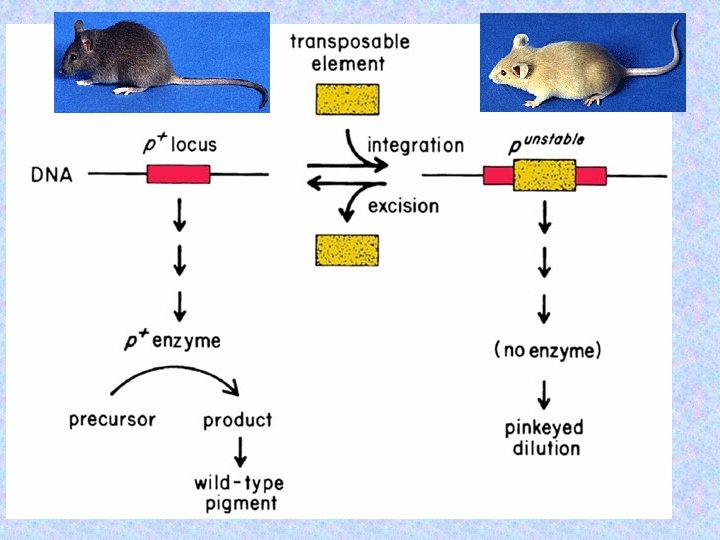
37

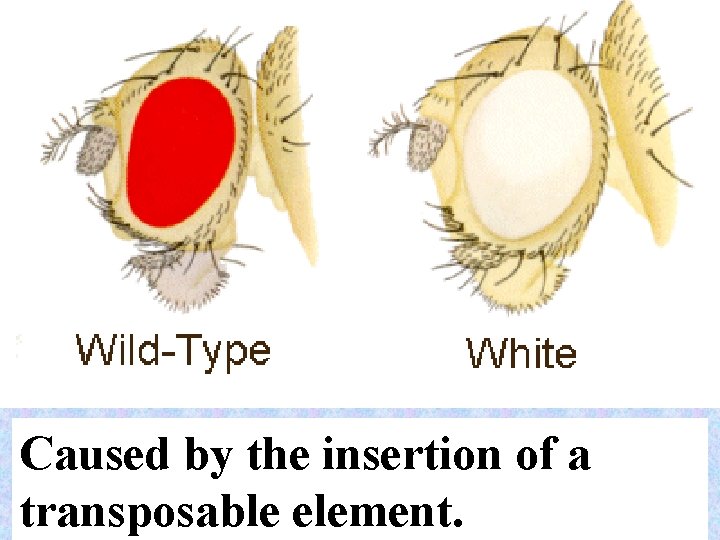
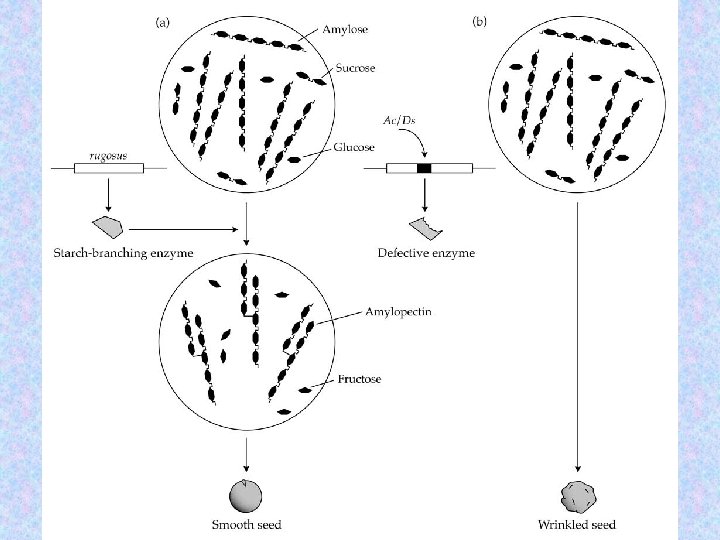
Caused by the insertion of a transposable element. 38

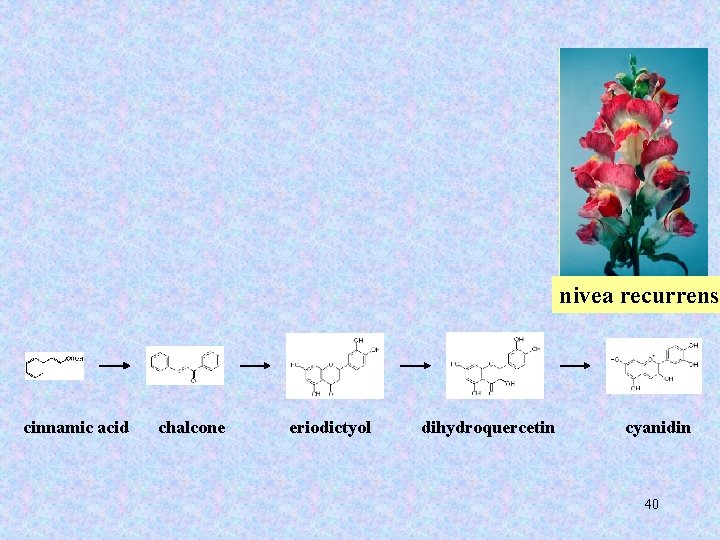
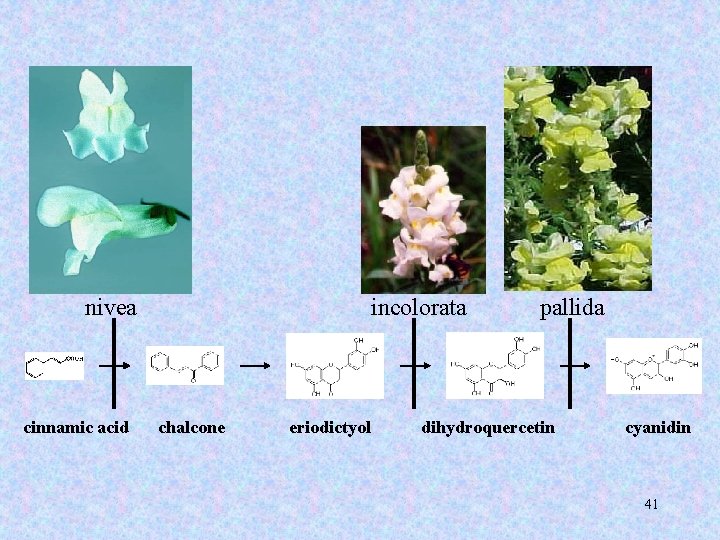
Transposable elements in Antirrhinum majus 39

nivea recurrens cinnamic acid chalcone eriodictyol dihydroquercetin cyanidin 40

nivea cinnamic acid incolorata chalcone eriodictyol pallida dihydroquercetin cyanidin 41

42

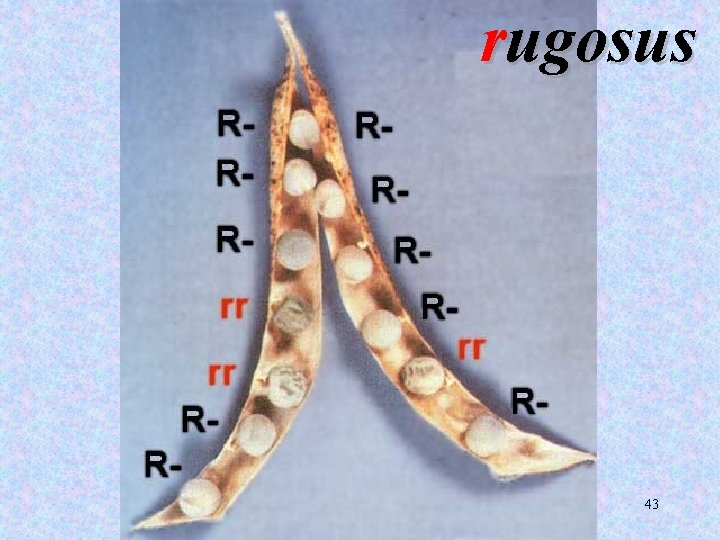
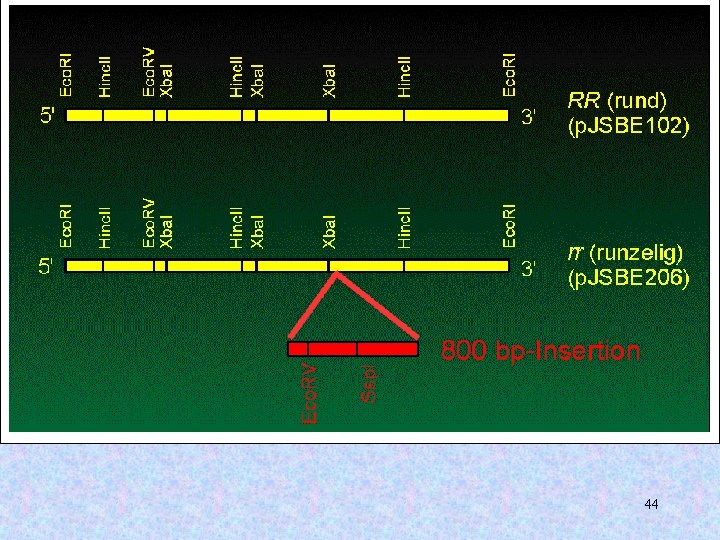
rugosus 43

44

45

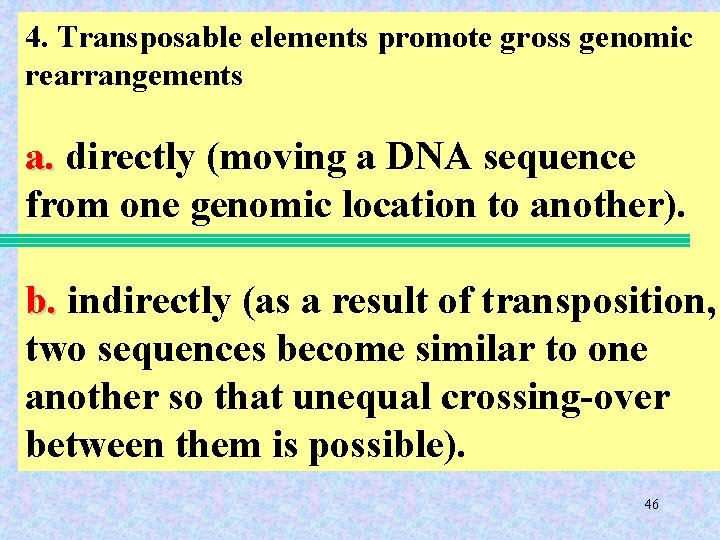
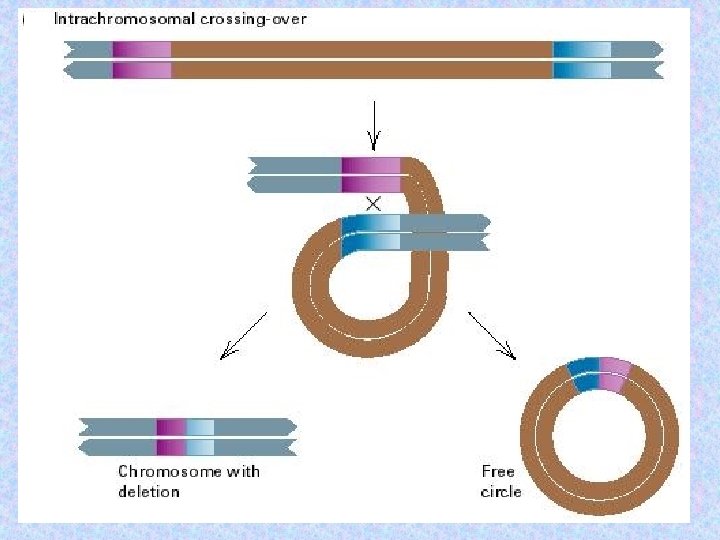
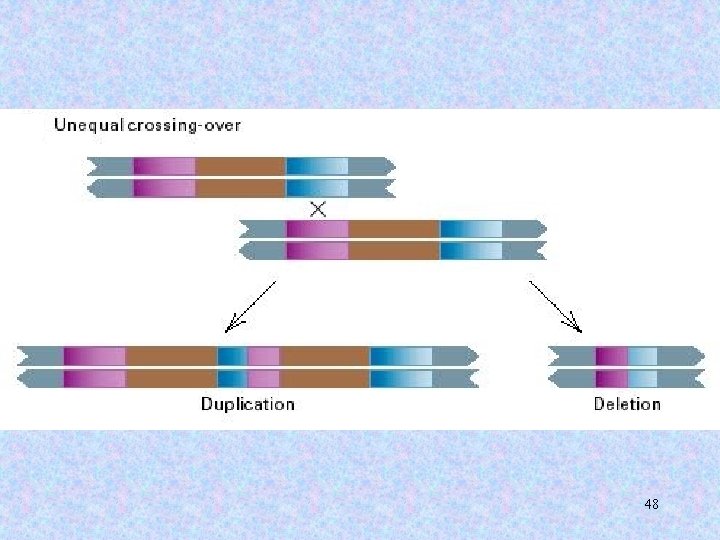
4. Transposable elements promote gross genomic rearrangements a. directly (moving a DNA sequence from one genomic location to another). b. indirectly (as a result of transposition, two sequences become similar to one another so that unequal crossing-over between them is possible). 46

47

48

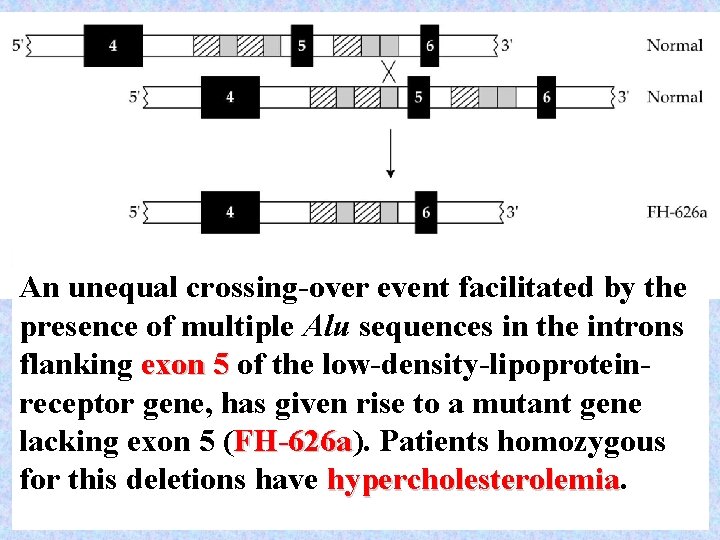
An unequal crossing-over event facilitated by the presence of multiple Alu sequences in the introns flanking exon 5 of the low-density-lipoproteinreceptor gene, has given rise to a mutant gene lacking exon 5 (FH-626 a). FH-626 a Patients homozygous for this deletions have hypercholesterolemia 49

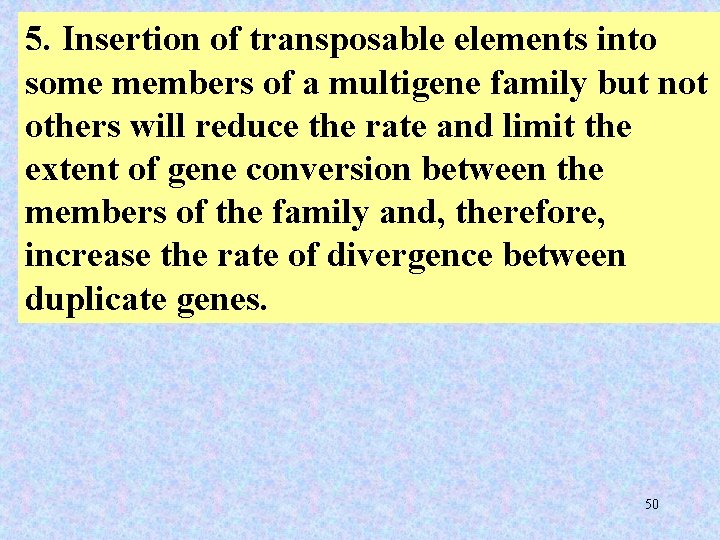
5. Insertion of transposable elements into some members of a multigene family but not others will reduce the rate and limit the extent of gene conversion between the members of the family and, therefore, increase the rate of divergence between duplicate genes. 50

6. Some transposable elements cause an increase in the rate of mutation in adjacent regions. 51

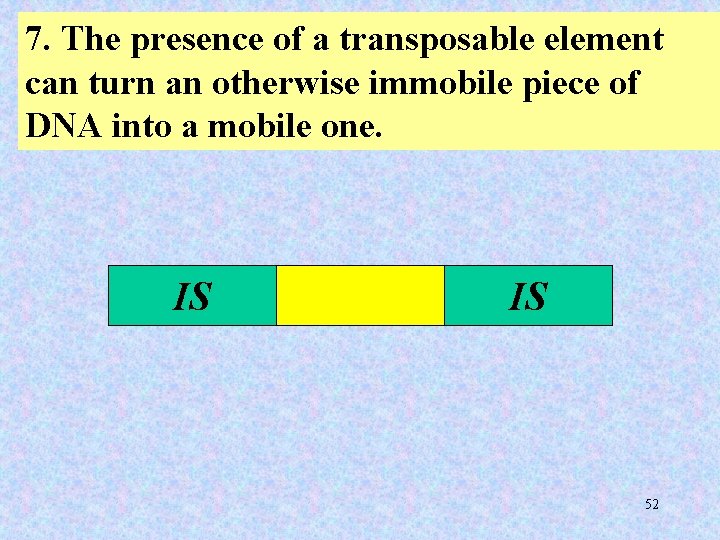
7. The presence of a transposable element can turn an otherwise immobile piece of DNA into a mobile one. IS IS 52

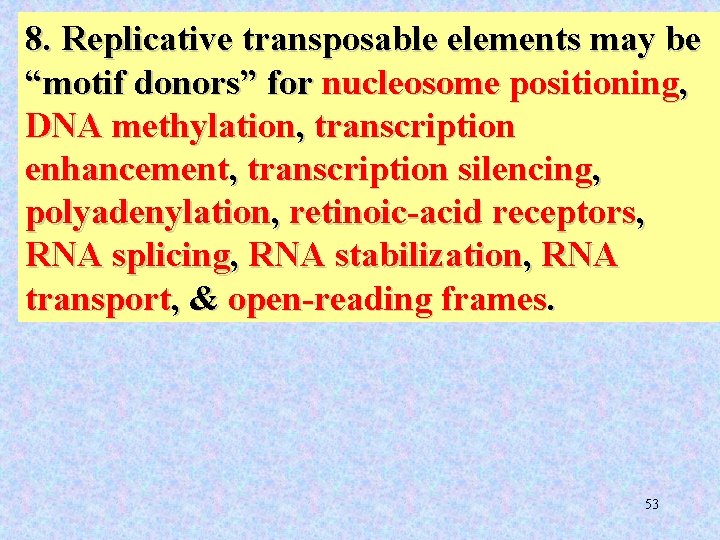
8. Replicative transposable elements may be “motif donors” for nucleosome positioning, DNA methylation, transcription enhancement, transcription silencing, polyadenylation, retinoic-acid receptors, RNA splicing, RNA stabilization, RNA transport, & open-reading frames. 53

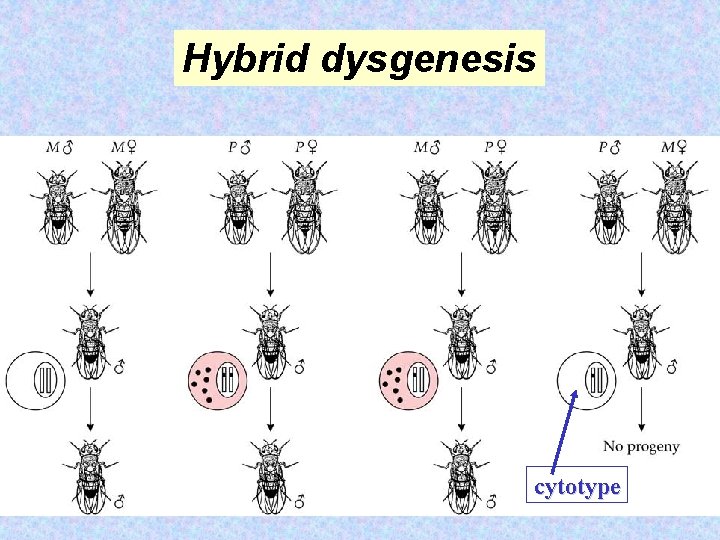
Hybrid dysgenesis in Drosophila is a syndrome of correlated abnormal genetic traits that is spontaneously induced in one type of hybrid between certain mutually interactive strains, but not in the reciprocal hybrid. Margaret Kidwell 54

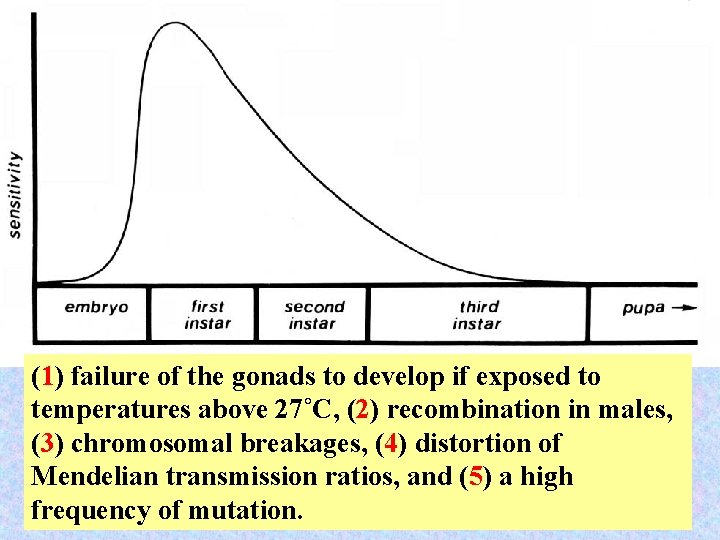
(1) failure of the gonads to develop if exposed to temperatures above 27˚C, (2) recombination in males, (3) chromosomal breakages, (4) distortion of Mendelian transmission ratios, and (5) a high 55 frequency of mutation.

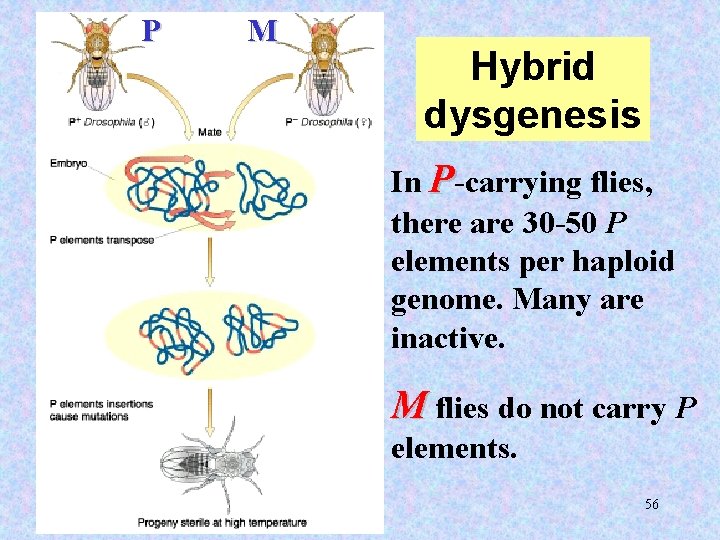
P M Hybrid dysgenesis In P-carrying flies, there are 30 -50 P elements per haploid genome. Many are inactive. M flies do not carry P elements. 56

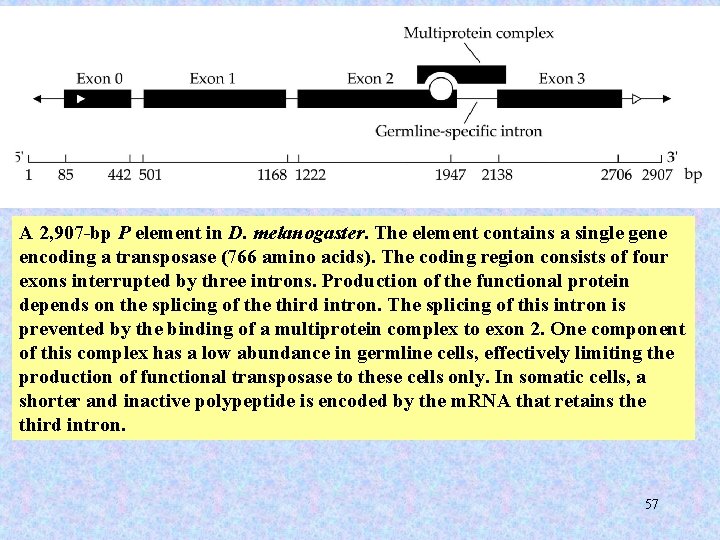
A 2, 907 -bp P element in D. melanogaster. The element contains a single gene encoding a transposase (766 amino acids). The coding region consists of four exons interrupted by three introns. Production of the functional protein depends on the splicing of the third intron. The splicing of this intron is prevented by the binding of a multiprotein complex to exon 2. One component of this complex has a low abundance in germline cells, effectively limiting the production of functional transposase to these cells only. In somatic cells, a shorter and inactive polypeptide is encoded by the m. RNA that retains the third intron. 57

Hybrid dysgenesis cytotype 58

Speciation (cladogenesis) = the creation of two or more taxa from a parental taxon. 59

60

Why NOT hybrid dysgenesis? Because of segregation distortion, P quickly spreads through the population. P has the ability to move from individual to individual as an infectious agent. Hybrid dysgenesis was only found in Drosophila mealnogaster and may not be a universal phenomenon. 61

“The change from species to species is not a change involving more and more additional atomistic changes, but a complete change of the primary pattern or reaction system into a new one, which afterwards may again produce intraspecific variation by micromutation. ” (Richard Goldschmidt. 1940. The Material Basis of Evolution, pp. 205 -206) Richard Goldschmidt The “hopeful monster” idea 62

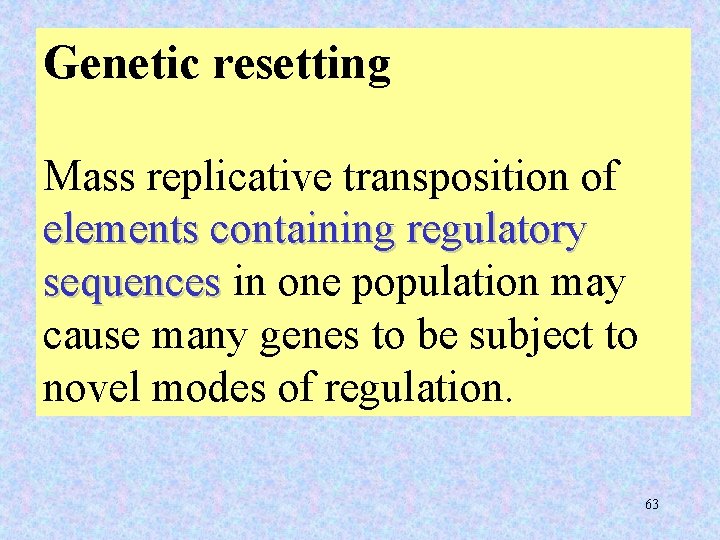
Genetic resetting Mass replicative transposition of elements containing regulatory sequences in one population may cause many genes to be subject to novel modes of regulation. 63

A genetically reset population will become reproductively isolated from the population that retains the old form of gene regulation. 64

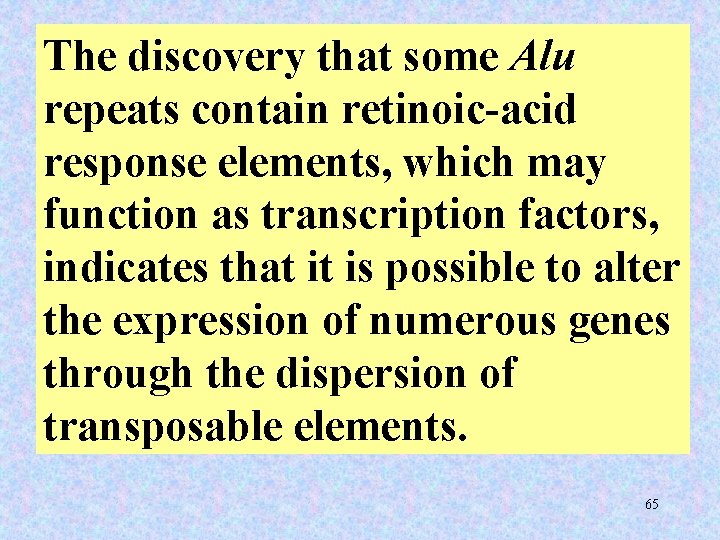
The discovery that some Alu repeats contain retinoic-acid response elements, which may function as transcription factors, indicates that it is possible to alter the expression of numerous genes through the dispersion of transposable elements. 65

Mechanical incompatibility In one population the transposable elements multiply to such an extent as to cause a significant increase in the size of the chromosomes. A hybrid organism that inherits big chromosomes from one parent and small chromosomes from the other would experience difficulties in chromosome pairing during meiosis, and would be most probably be sterile. 66

Thursday, April 26, 2012 Tuesday, May 8, 2012 final exam 67