Linear Models for Classification Probabilistic Methods Adopted from
Linear Models for Classification: Probabilistic Methods Adopted from Seung-Joon Yi Biointelligence Laboratory, Seoul National University http: //bi. snu. ac. kr/

Recall, Linear Methods for Classification Problem Definition: Given the training data {xn, tn}, find a linear model for each class yk(x) to partition the feature space into decision regions l Deterministic Models: ¨ Discriminant Functions ¨ Fisher Discriminant function ¨ Perceptron 2
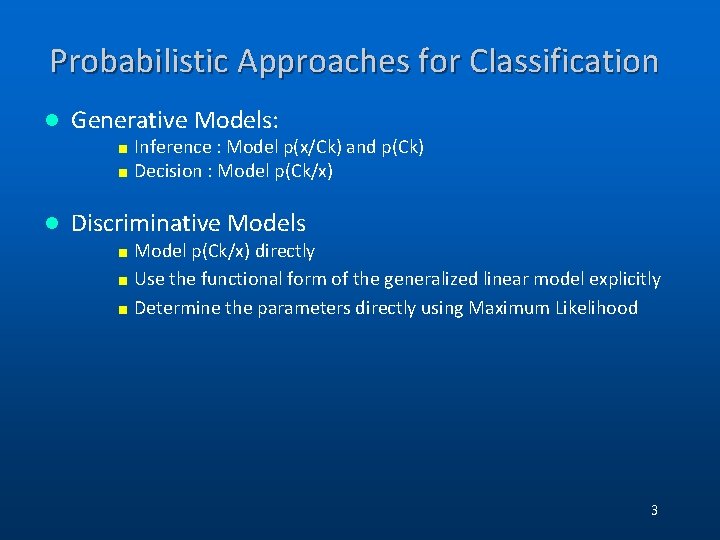
Probabilistic Approaches for Classification l Generative Models: < Inference : Model p(x/Ck) and p(Ck) < Decision : Model p(Ck/x) l Discriminative Models < Model p(Ck/x) directly < Use the functional form of the generalized linear model explicitly < Determine the parameters directly using Maximum Likelihood 3
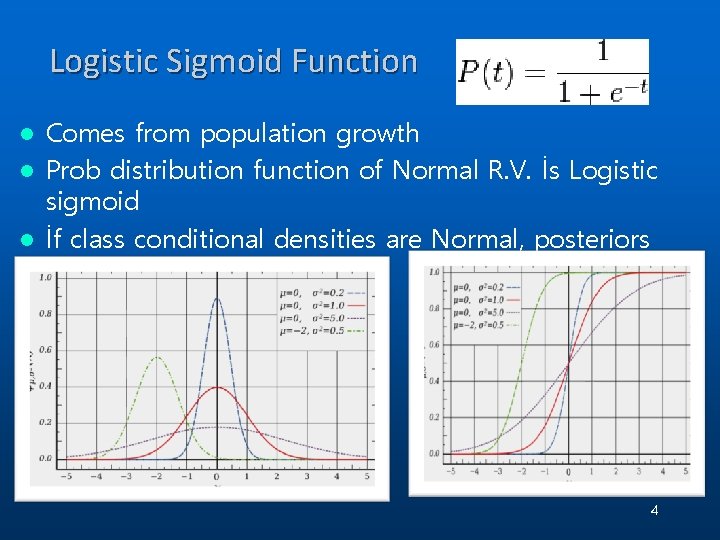
Logistic Sigmoid Function Comes from population growth l Prob distribution function of Normal R. V. İs Logistic sigmoid l İf class conditional densities are Normal, posteriors become logistic sigmoid l 4

Posterior Probabilities can be formulated by 2 -Class: Logistic sigmoid acting on a linear function of x l K-Class: Softmax transformation of a linear function of x l Then, l The parameters of the densities as well as the class priors can be determined using Maximum Likelihood 5
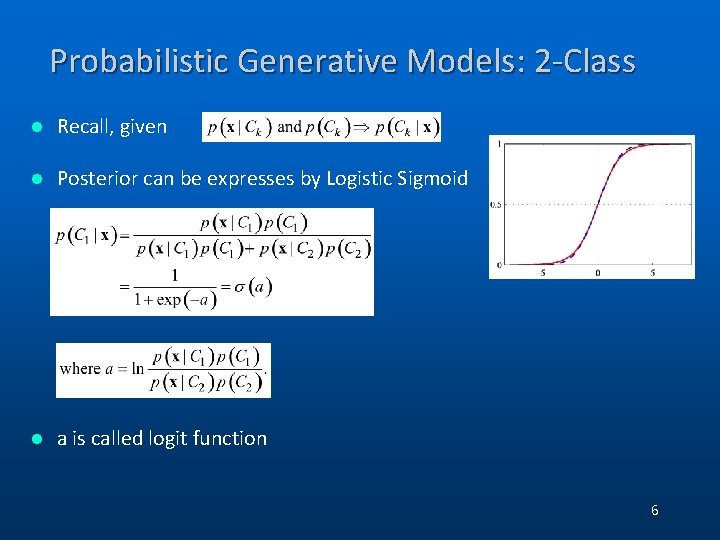
Probabilistic Generative Models: 2 -Class l Recall, given l Posterior can be expresses by Logistic Sigmoid l a is called logit function 6
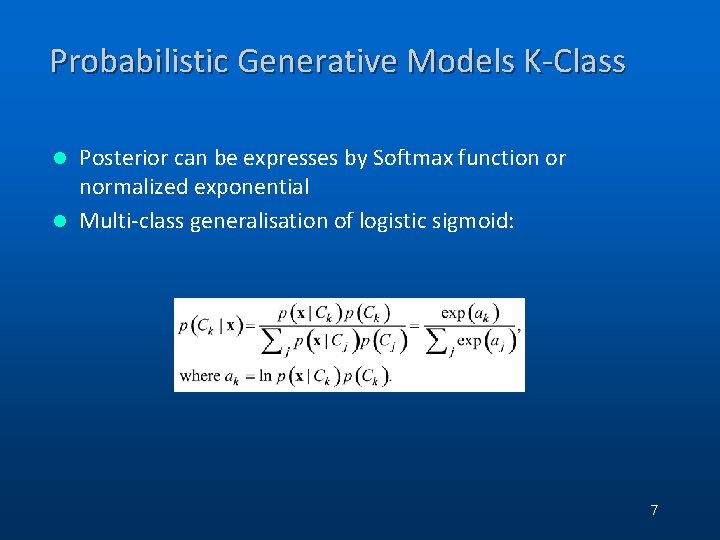
Probabilistic Generative Models K-Class Posterior can be expresses by Softmax function or normalized exponential l Multi-class generalisation of logistic sigmoid: l 7
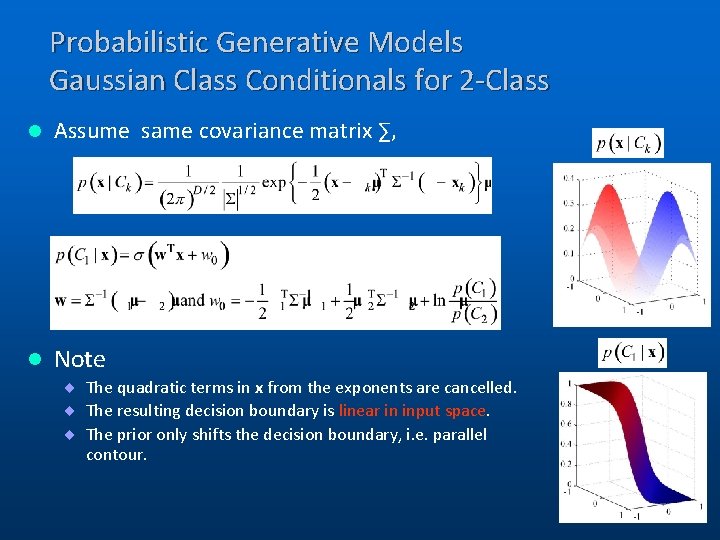
Probabilistic Generative Models Gaussian Class Conditionals for 2 -Class l Assume same covariance matrix ∑, l Note ¨ The quadratic terms in x from the exponents are cancelled. ¨ The resulting decision boundary is linear in input space. ¨ The prior only shifts the decision boundary, i. e. parallel contour. 8
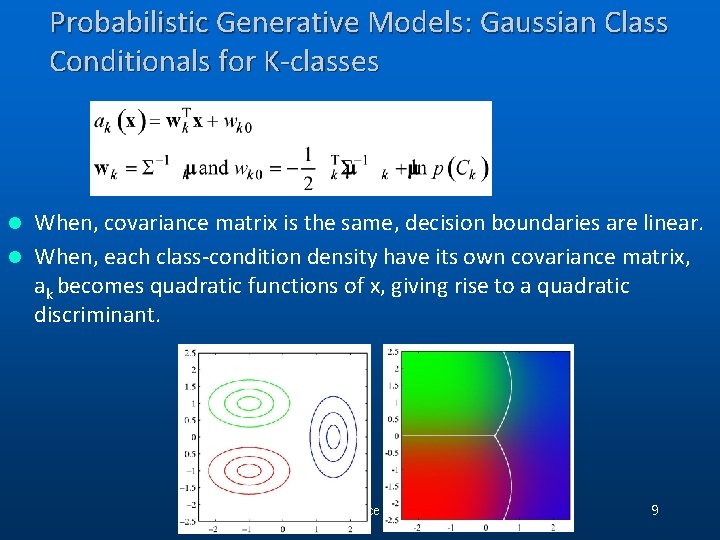
Probabilistic Generative Models: Gaussian Class Conditionals for K-classes When, covariance matrix is the same, decision boundaries are linear. l When, each class-condition density have its own covariance matrix, ak becomes quadratic functions of x, giving rise to a quadratic discriminant. l (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 9
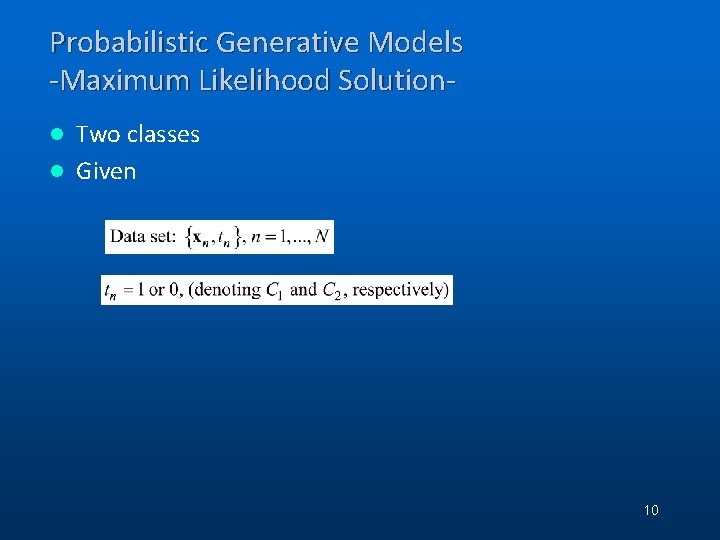
Probabilistic Generative Models -Maximum Likelihood Solution. Two classes l Given l 10

Q: Find P(C 1) = π and P(C 2) = 1 - π and parameters of p(Ck/x): μ 1, μ 2 and 11
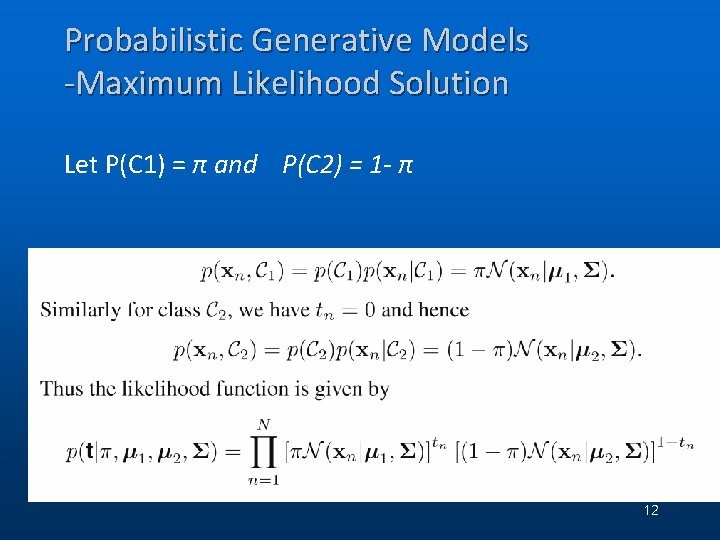
Probabilistic Generative Models -Maximum Likelihood Solution Let P(C 1) = π and P(C 2) = 1 - π 12
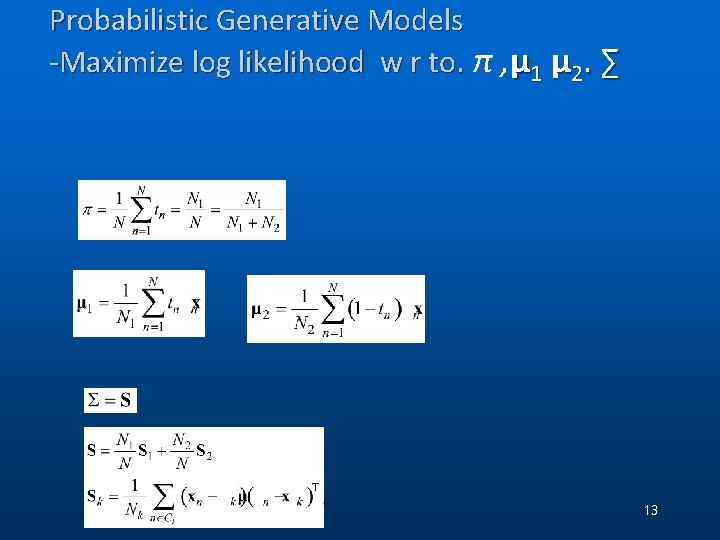
Probabilistic Generative Models -Maximize log likelihood w r to. π , μ 1 μ 2. ∑ . 13
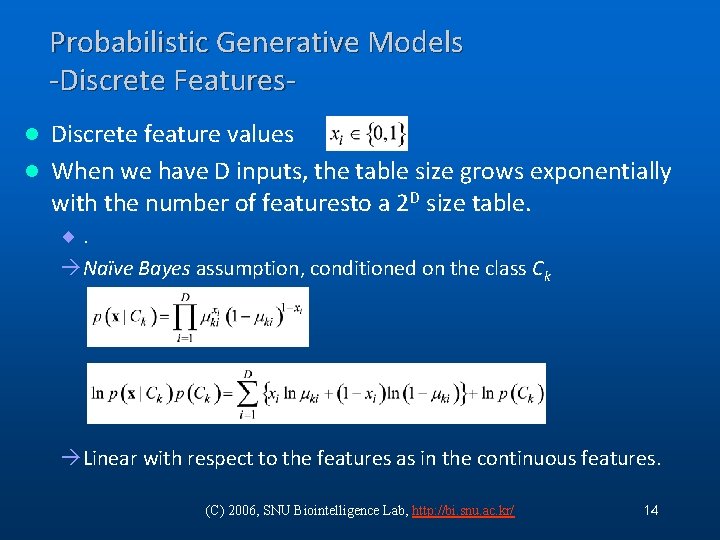
Probabilistic Generative Models -Discrete Features. Discrete feature values l When we have D inputs, the table size grows exponentially with the number of featuresto a 2 D size table. l ¨. àNaïve Bayes assumption, conditioned on the class Ck àLinear with respect to the features as in the continuous features. (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 14
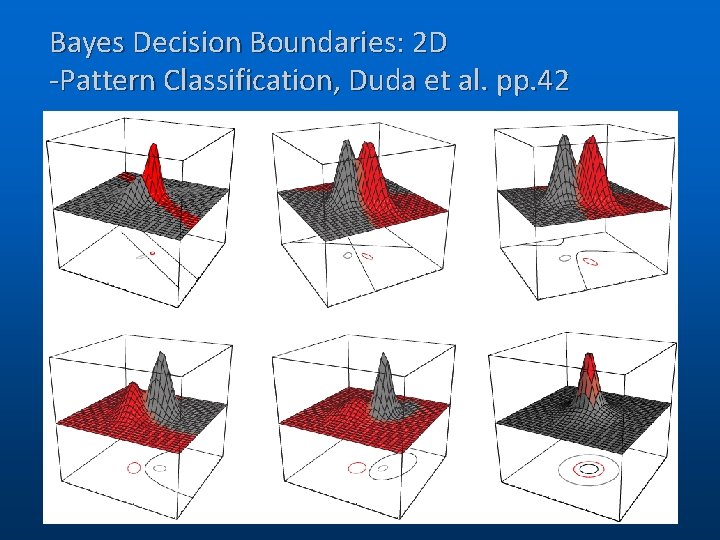
Bayes Decision Boundaries: 2 D -Pattern Classification, Duda et al. pp. 42 (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 15
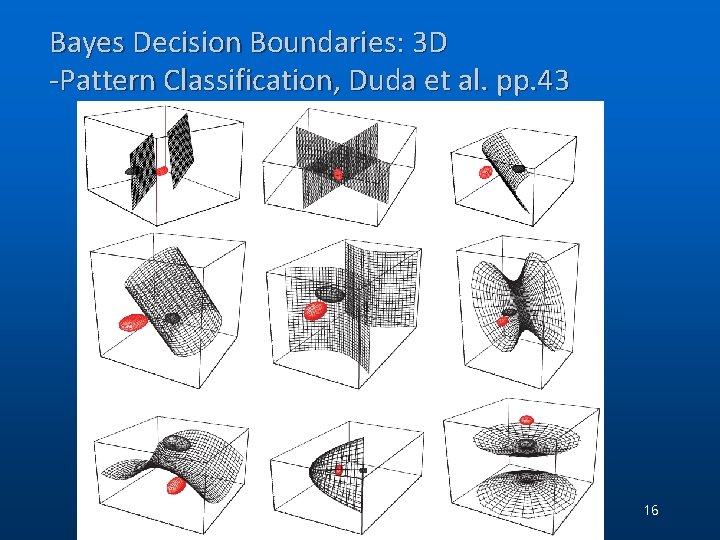
Bayes Decision Boundaries: 3 D -Pattern Classification, Duda et al. pp. 43 (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 16

For both Gaussian distributed and discrete inputs l The posterior class probabilities are given by ¨ Generalized linear models with logistic sigmoid or ¨ softmax activation functions. 17
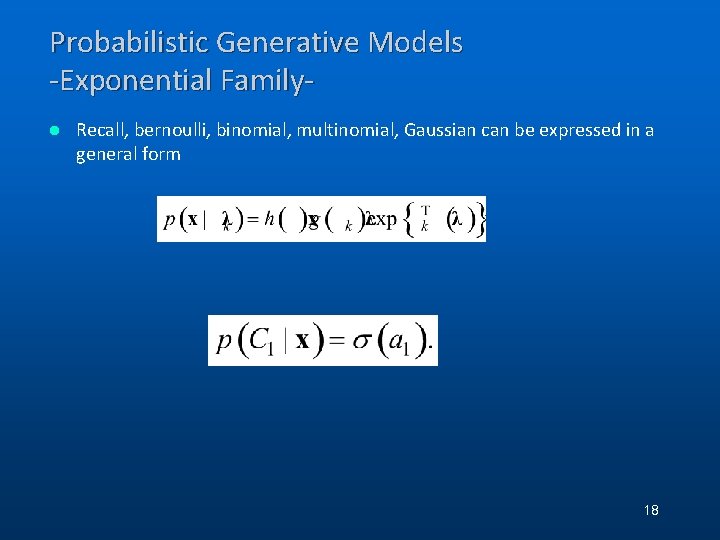
Probabilistic Generative Models -Exponential Familyl Recall, bernoulli, binomial, multinomial, Gaussian can be expressed in a general form 18
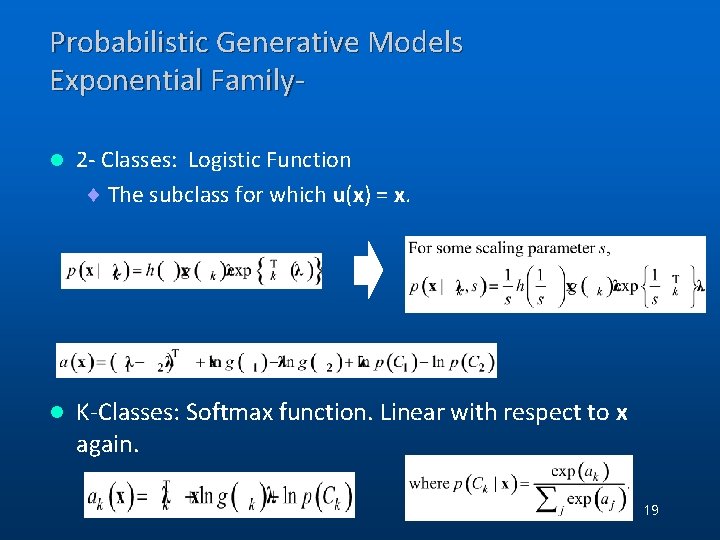
Probabilistic Generative Models Exponential Familyl 2 - Classes: Logistic Function ¨ The subclass for which u(x) = x. l K-Classes: Softmax function. Linear with respect to x again. 19

Probabilistic Discriminative Models Goal: Find p(Ck/x) directly l No inferrence step l Discriminative Training: Max likelihood p(Ck/x) l İmproves prediction performance when p(x/Ck) is poorly estimated l 20
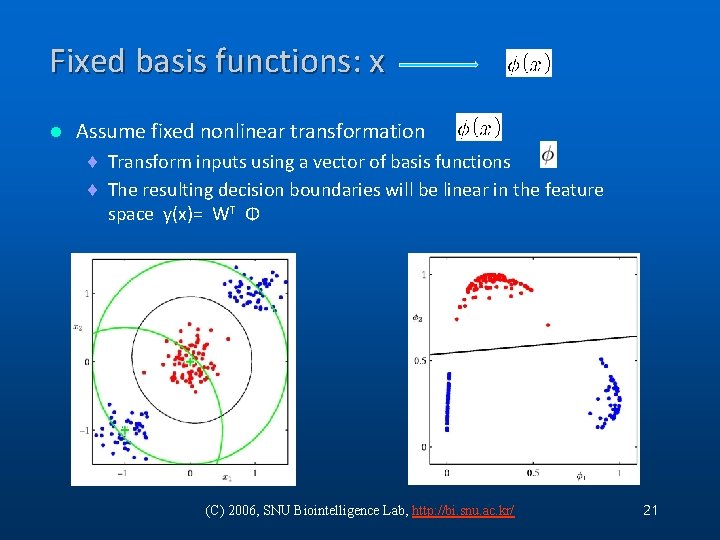
Fixed basis functions: x l Assume fixed nonlinear transformation ¨ Transform inputs using a vector of basis functions ¨ The resulting decision boundaries will be linear in the feature space y(x)= WT Φ (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 21
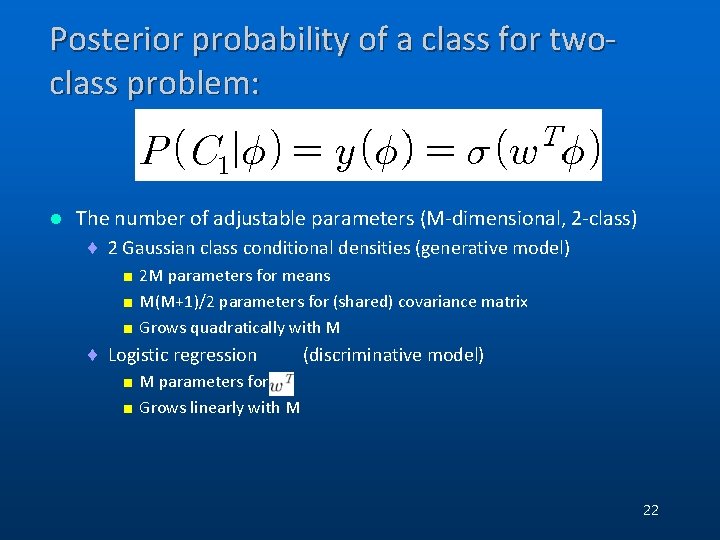
Posterior probability of a class for twoclass problem: l The number of adjustable parameters (M-dimensional, 2 -class) ¨ 2 Gaussian class conditional densities (generative model) < 2 M parameters for means < M(M+1)/2 parameters for (shared) covariance matrix < Grows quadratically with M ¨ Logistic regression (discriminative model) < M parameters for < Grows linearly with M 22
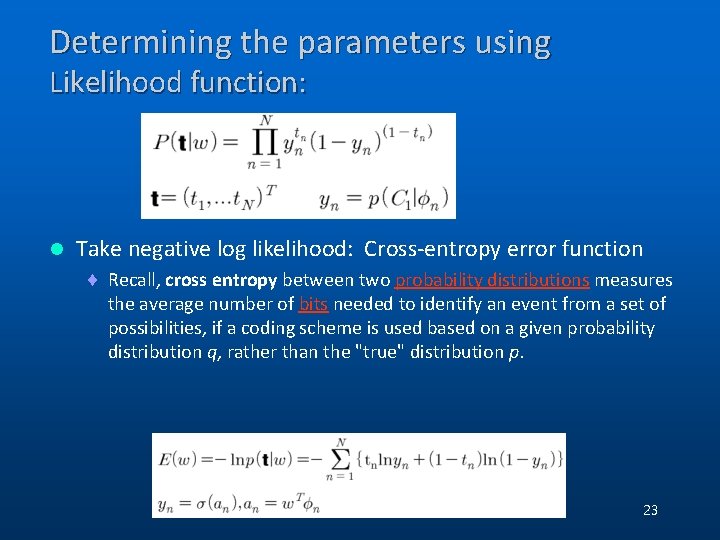
Determining the parameters using Likelihood function: l Take negative log likelihood: Cross-entropy error function ¨ Recall, cross entropy between two probability distributions measures the average number of bits needed to identify an event from a set of possibilities, if a coding scheme is used based on a given probability distribution q, rather than the "true" distribution p. (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 23
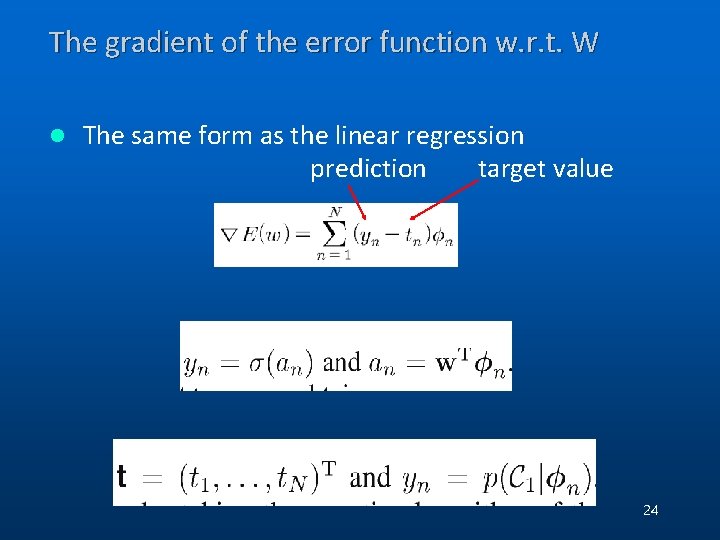
The gradient of the error function w. r. t. W l The same form as the linear regression prediction target value 24
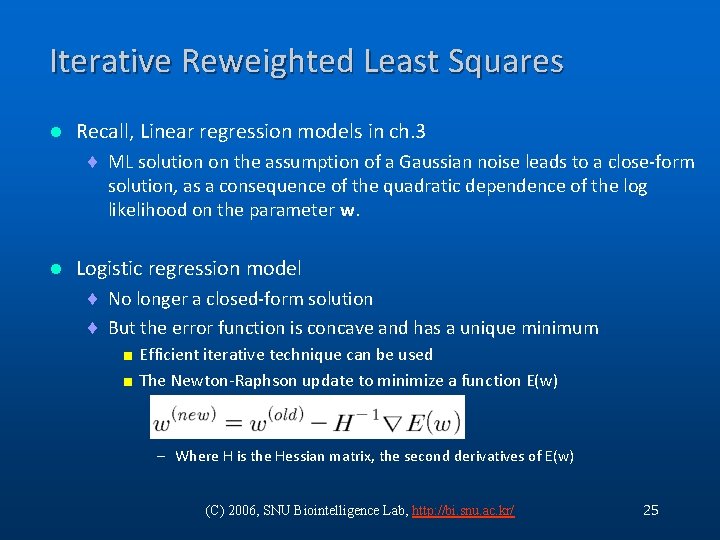
Iterative Reweighted Least Squares l Recall, Linear regression models in ch. 3 ¨ ML solution on the assumption of a Gaussian noise leads to a close-form solution, as a consequence of the quadratic dependence of the log likelihood on the parameter w. l Logistic regression model ¨ No longer a closed-form solution ¨ But the error function is concave and has a unique minimum < Efficient iterative technique can be used < The Newton-Raphson update to minimize a function E(w) – Where H is the Hessian matrix, the second derivatives of E(w) (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 25
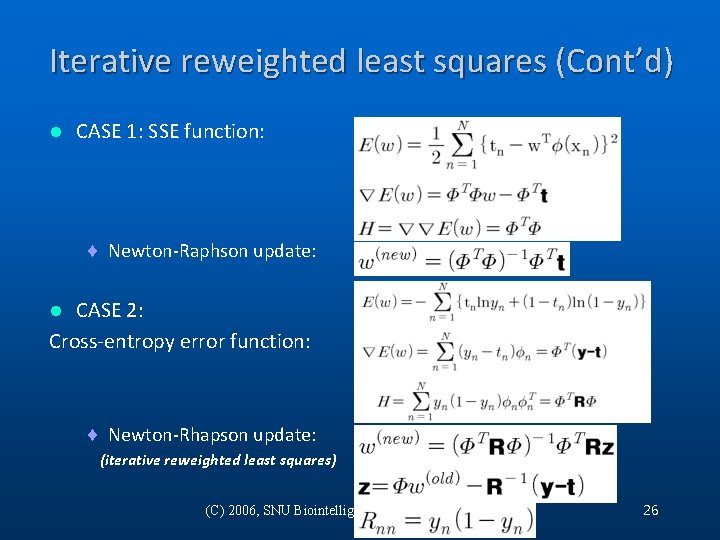
Iterative reweighted least squares (Cont’d) l CASE 1: SSE function: ¨ Newton-Raphson update: CASE 2: Cross-entropy error function: l ¨ Newton-Rhapson update: (iterative reweighted least squares) (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 26
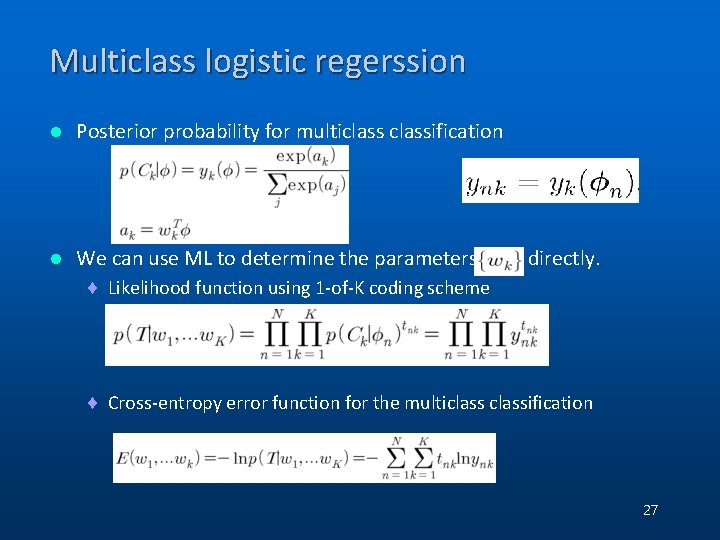
Multiclass logistic regerssion l Posterior probability for multiclassification l We can use ML to determine the parameters directly. ¨ Likelihood function using 1 -of-K coding scheme ¨ Cross-entropy error function for the multiclassification 27
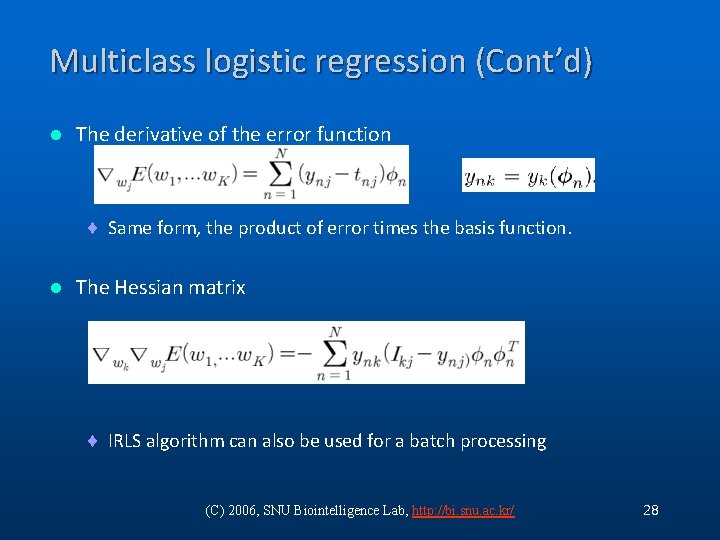
Multiclass logistic regression (Cont’d) l The derivative of the error function ¨ Same form, the product of error times the basis function. l The Hessian matrix ¨ IRLS algorithm can also be used for a batch processing (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 28
Generalized Linear Models l Recall, for a broad range of class-conditional distributions, described by the exponential family, the resulting posterior class probabilities are given by a logistic(or softmax) transformation acting on a linear function of the feature variables. l However this is not the case for all choices of class-conditional density ¨ It might be worth exploring other types of discriminative probabilistic model (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 29
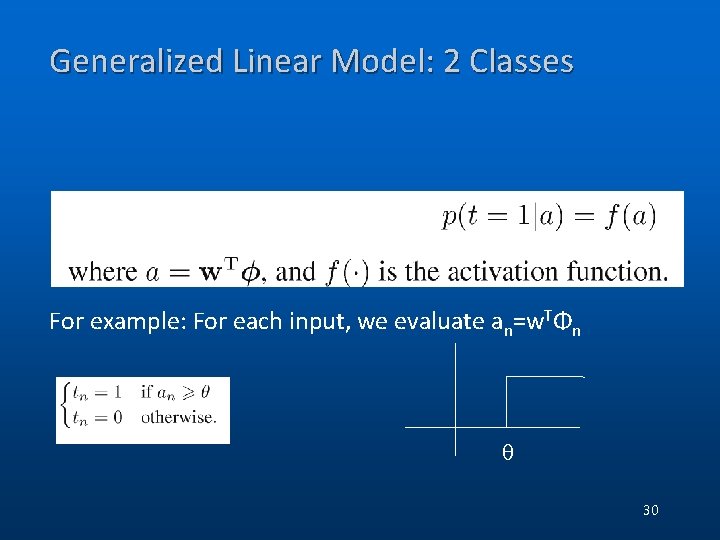
Generalized Linear Model: 2 Classes For example: For each input, we evaluate an=w. TΦn θ 30
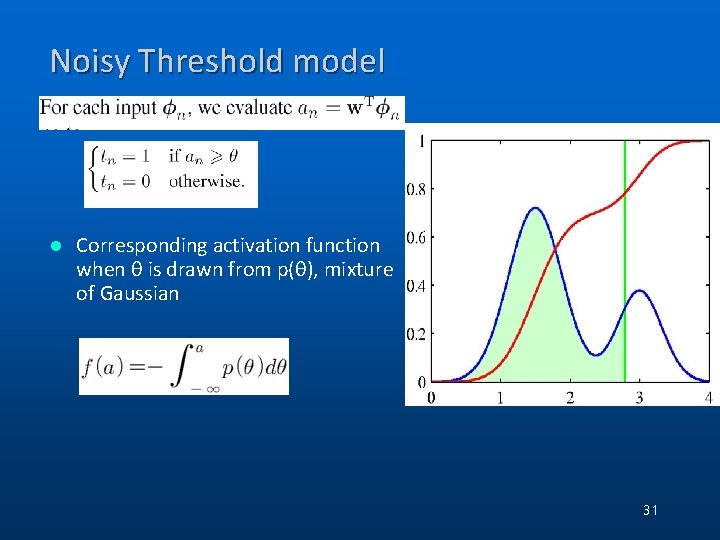
Noisy Threshold model l Corresponding activation function when θ is drawn from p(θ), mixture of Gaussian 31
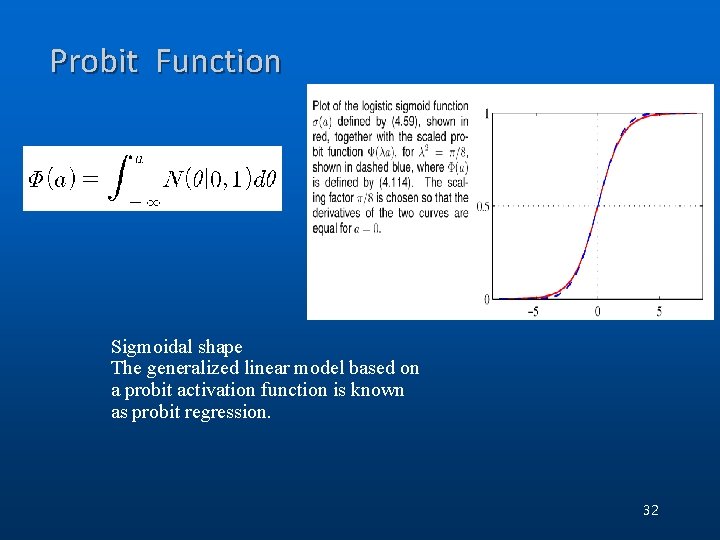
Probit Function Sigmoidal shape The generalized linear model based on a probit activation function is known as probit regression. 32
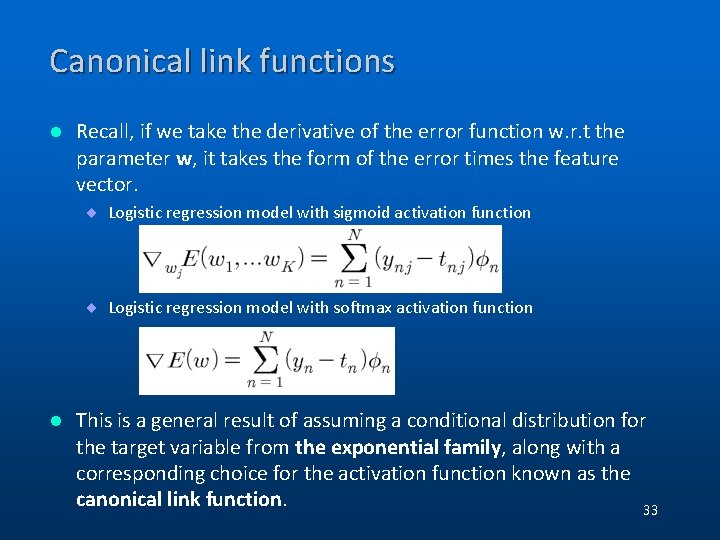
Canonical link functions l Recall, if we take the derivative of the error function w. r. t the parameter w, it takes the form of the error times the feature vector. ¨ Logistic regression model with sigmoid activation function ¨ Logistic regression model with softmax activation function l This is a general result of assuming a conditional distribution for the target variable from the exponential family, along with a corresponding choice for the activation function known as the canonical link function. 33
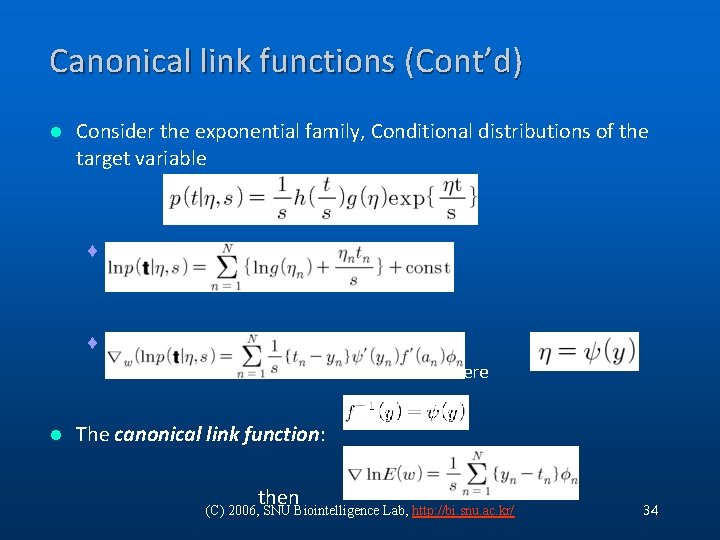
Canonical link functions (Cont’d) l Consider the exponential family, Conditional distributions of the target variable ¨ Log likelihood: ¨ The derivative of the log likelihood: where l The canonical link function: then (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 34
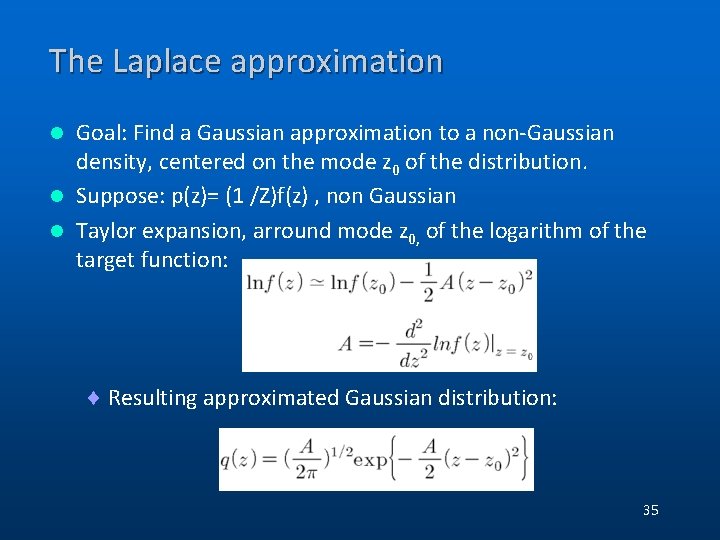
The Laplace approximation Goal: Find a Gaussian approximation to a non-Gaussian density, centered on the mode z 0 of the distribution. l Suppose: p(z)= (1 /Z)f(z) , non Gaussian l Taylor expansion, arround mode z 0, of the logarithm of the target function: l ¨ Resulting approximated Gaussian distribution: 35
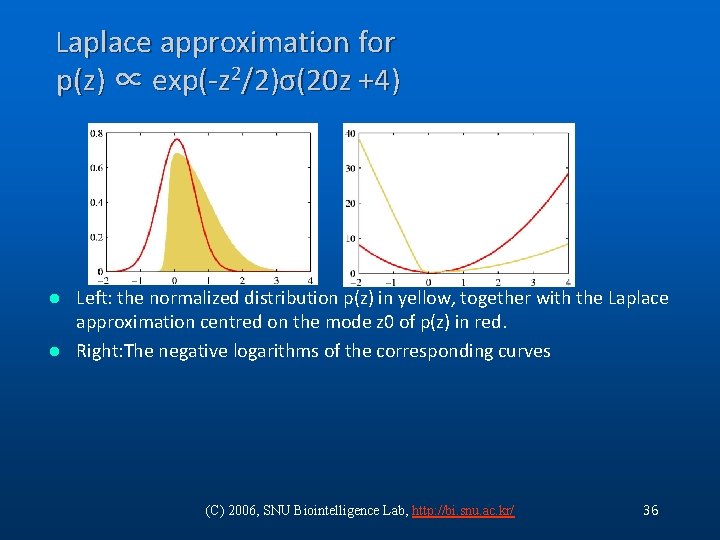
Laplace approximation for p(z) ∝ exp(-z 2/2)σ(20 z +4) Left: the normalized distribution p(z) in yellow, together with the Laplace approximation centred on the mode z 0 of p(z) in red. l Right: The negative logarithms of the corresponding curves l (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 36
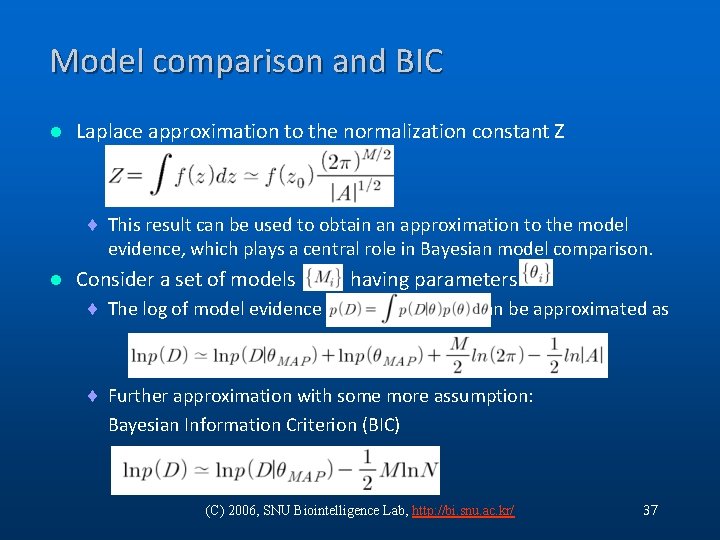
Model comparison and BIC l Laplace approximation to the normalization constant Z ¨ This result can be used to obtain an approximation to the model evidence, which plays a central role in Bayesian model comparison. l Consider a set of models ¨ The log of model evidence having parameters can be approximated as ¨ Further approximation with some more assumption: Bayesian Information Criterion (BIC) (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 37
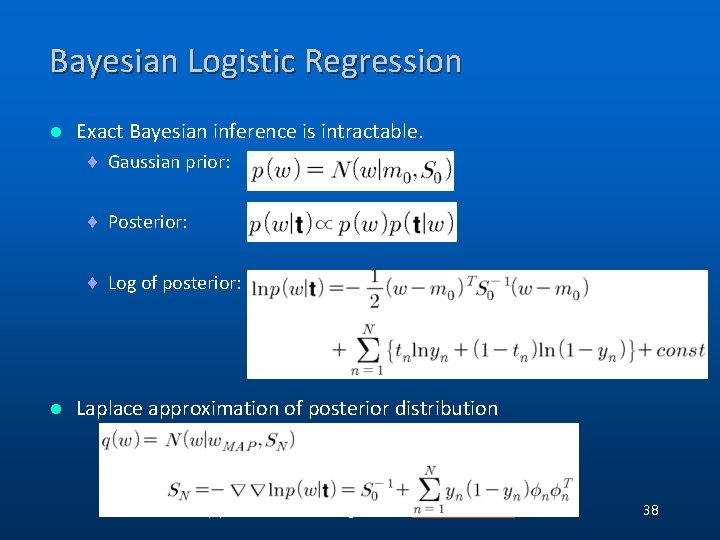
Bayesian Logistic Regression l Exact Bayesian inference is intractable. ¨ Gaussian prior: ¨ Posterior: ¨ Log of posterior: l Laplace approximation of posterior distribution (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 38
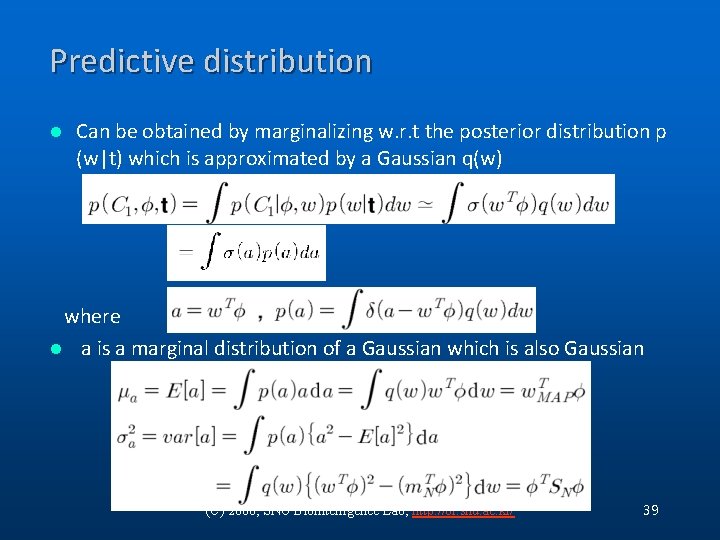
Predictive distribution l Can be obtained by marginalizing w. r. t the posterior distribution p (w|t) which is approximated by a Gaussian q(w) where l a is a marginal distribution of a Gaussian which is also Gaussian (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 39
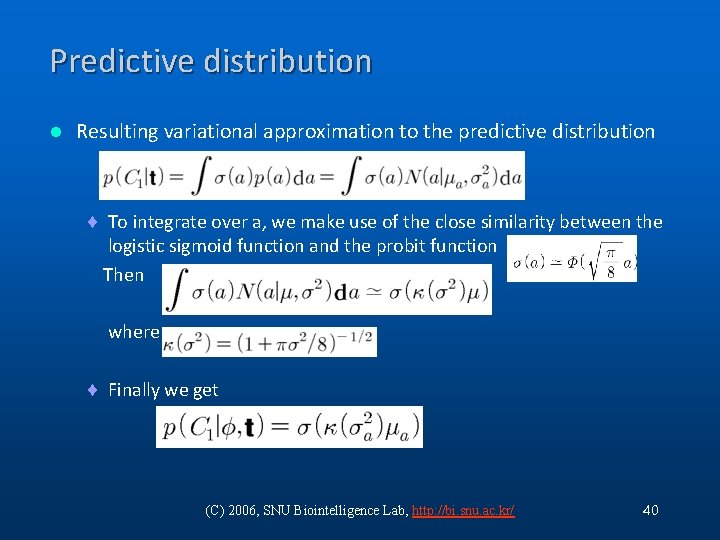
Predictive distribution l Resulting variational approximation to the predictive distribution ¨ To integrate over a, we make use of the close similarity between the logistic sigmoid function and the probit function Then where ¨ Finally we get (C) 2006, SNU Biointelligence Lab, http: //bi. snu. ac. kr/ 40
- Slides: 40