Leveraging EST Sequencing Micro Array Experiments and Database
Leveraging EST Sequencing, Micro Array Experiments and Database Integration for Gene Expression Analyses The Computational Biology and Informatics Laboratory http: //www. cbil. upenn. edu
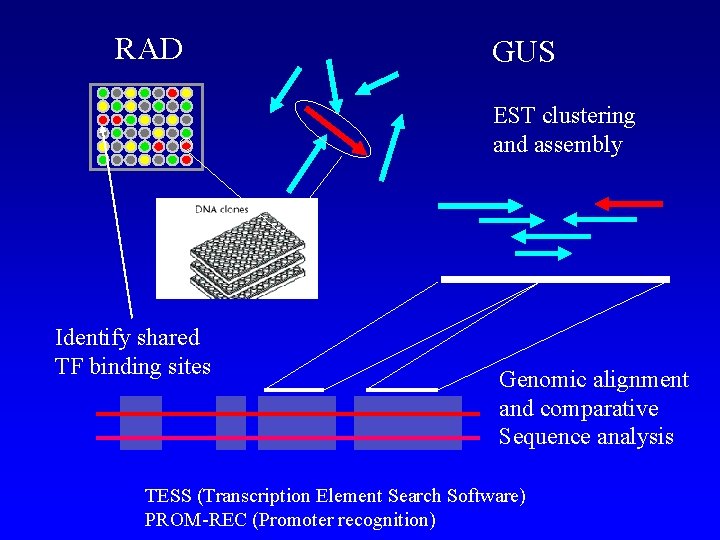
RAD GUS EST clustering and assembly Identify shared TF binding sites Genomic alignment and comparative Sequence analysis TESS (Transcription Element Search Software) PROM-REC (Promoter recognition)
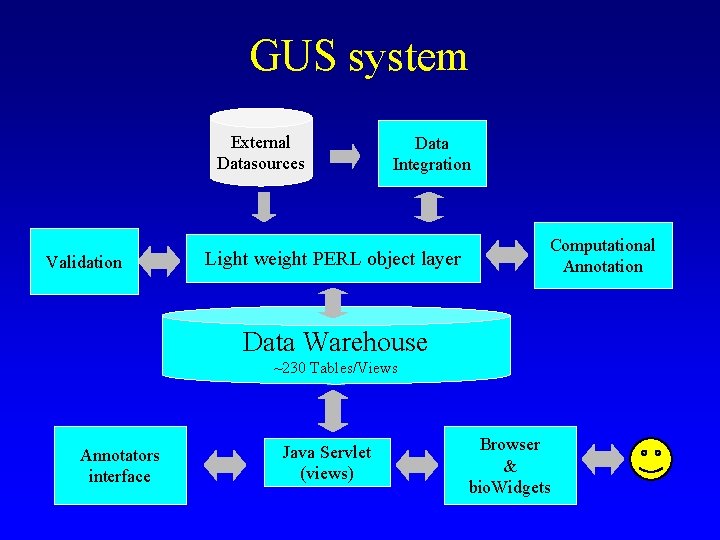
GUS system External Datasources Validation Data Integration Light weight PERL object layer Computational Annotation Data Warehouse ~230 Tables/Views Annotators interface Java Servlet (views) Browser & bio. Widgets
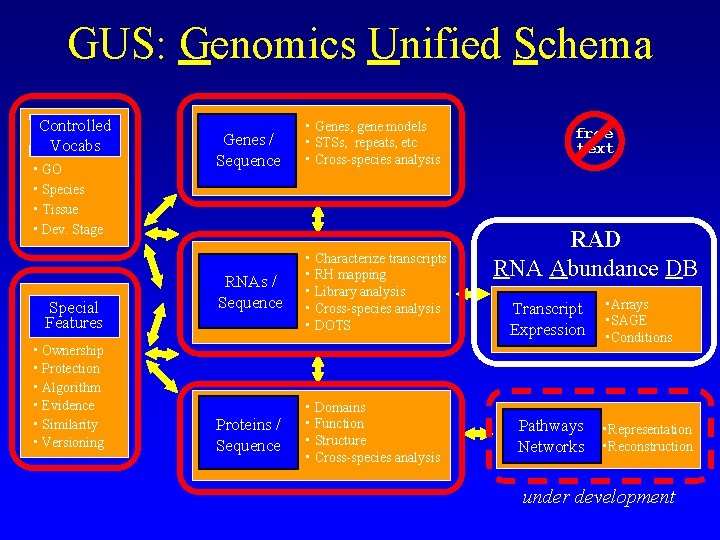
GUS: Genomics Unified Schema Controlled Vocabs • GO • Species • Tissue • Dev. Stage Special Features • Ownership • Protection • Algorithm • Evidence • Similarity • Versioning Genes / Sequence • Genes, gene models • STSs, repeats, etc • Cross-species analysis RNAs / Sequence • Characterize transcripts • RH mapping • Library analysis • Cross-species analysis • DOTS Proteins / Sequence • Domains • Function • Structure • Cross-species analysis free text RAD RNA Abundance DB Transcript Expression Pathways Networks • Arrays • SAGE • Conditions • Representation • Reconstruction under development
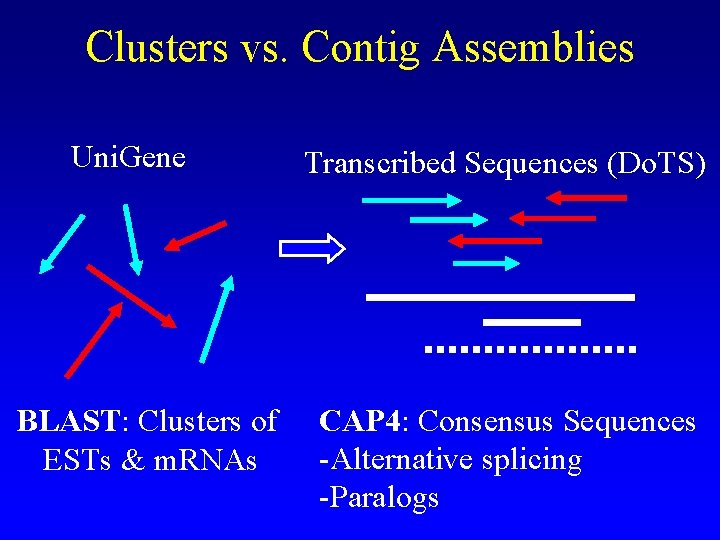
Clusters vs. Contig Assemblies Uni. Gene BLAST: Clusters of ESTs & m. RNAs Transcribed Sequences (Do. TS) CAP 4: Consensus Sequences -Alternative splicing -Paralogs
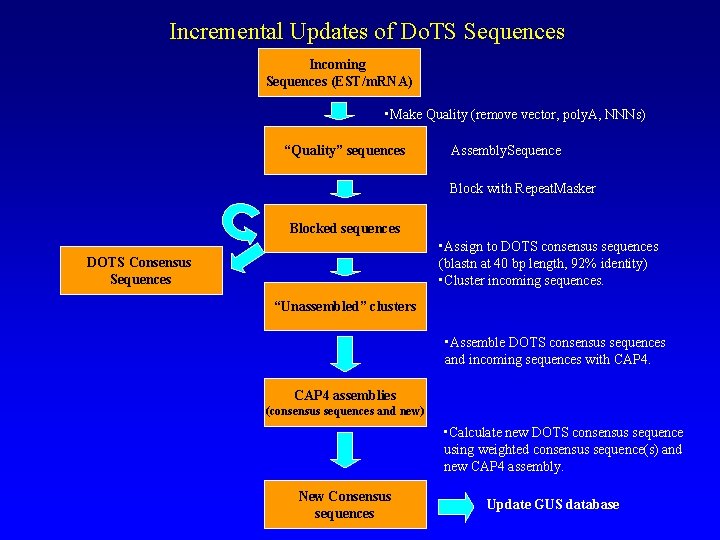
Incremental Updates of Do. TS Sequences Incoming Sequences (EST/m. RNA) • Make Quality (remove vector, poly. A, NNNs) “Quality” sequences Assembly. Sequence Block with Repeat. Masker Blocked sequences • Assign to DOTS consensus sequences (blastn at 40 bp length, 92% identity) • Cluster incoming sequences. DOTS Consensus Sequences “Unassembled” clusters • Assemble DOTS consensus sequences and incoming sequences with CAP 4 assemblies (consensus sequences and new) • Calculate new DOTS consensus sequence using weighted consensus sequence(s) and new CAP 4 assembly. New Consensus sequences Update GUS database
Assembly Validation • Alignment to Genomic Sequence via Blast/sim 4. • preliminary data look good • Assembly consistency (Assemblies provide potential SNPs)
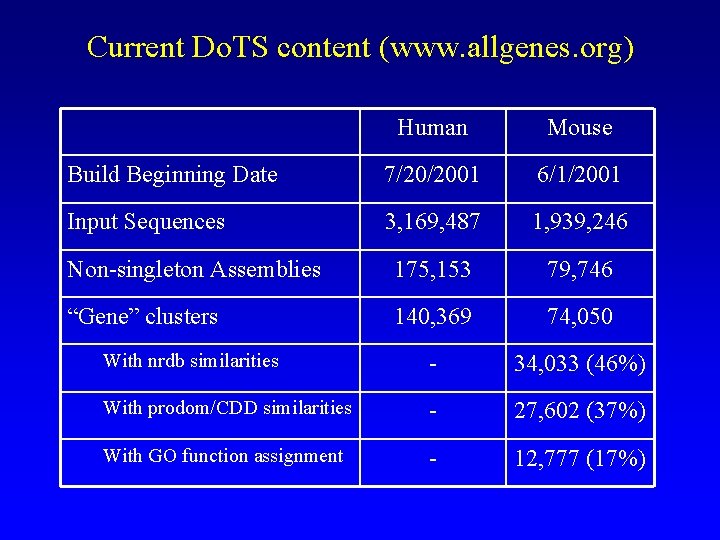
Current Do. TS content (www. allgenes. org) Human Mouse Build Beginning Date 7/20/2001 6/1/2001 Input Sequences 3, 169, 487 1, 939, 246 Non-singleton Assemblies 175, 153 79, 746 “Gene” clusters 140, 369 74, 050 With nrdb similarities - 34, 033 (46%) With prodom/CDD similarities - 27, 602 (37%) With GO function assignment - 12, 777 (17%)
Multiple labs Multiple biological systems Multiple platforms Expressed genes? Differentially-expressed genes? Co-regulated genes? Gene pathways?
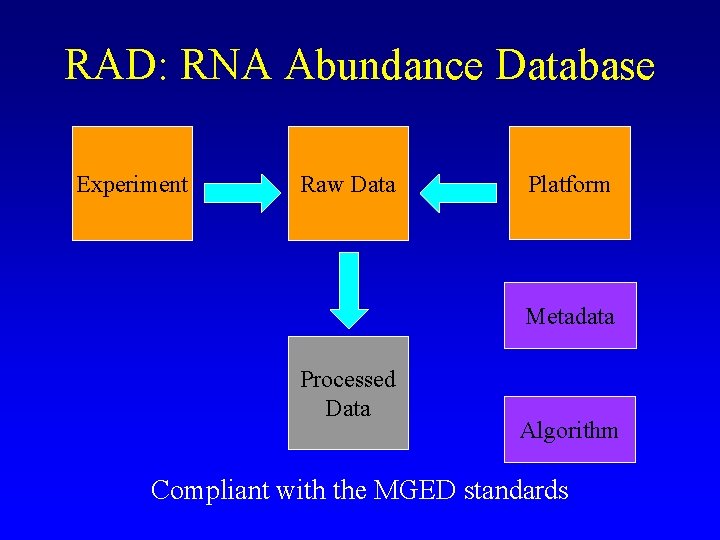
RAD: RNA Abundance Database Experiment Raw Data Platform Metadata Processed Data Algorithm Compliant with the MGED standards
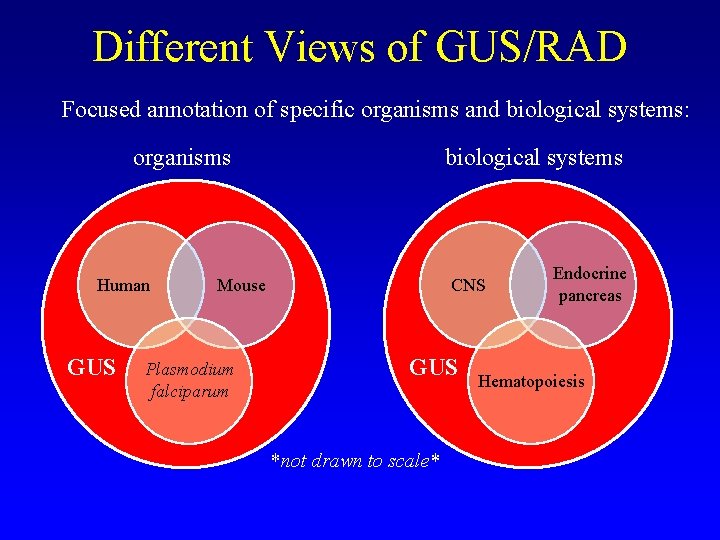
Different Views of GUS/RAD Focused annotation of specific organisms and biological systems: organisms Human GUS biological systems Mouse Plasmodium falciparum CNS GUS *not drawn to scale* Endocrine pancreas Hematopoiesis
WWW. CBIL. UPENN. EDU/EPCONDB
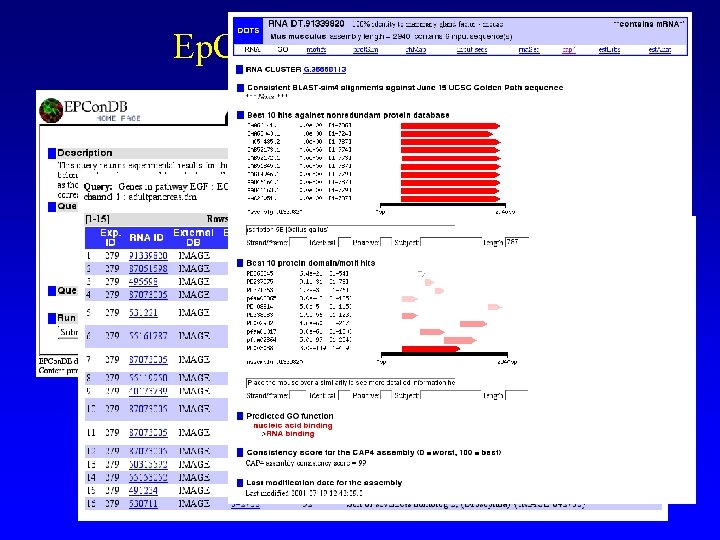
Ep. Con. DB Pathway query

WWW. PLASMODB. ORG
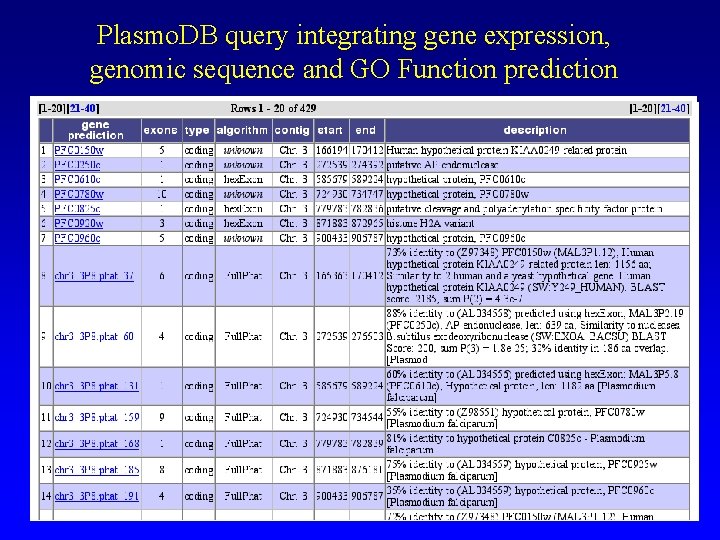
Plasmo. DB query integrating gene expression, genomic sequence and GO Function prediction
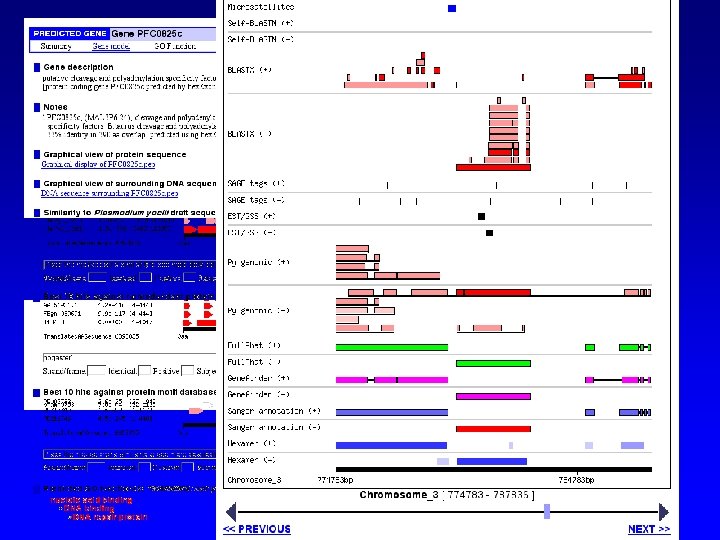
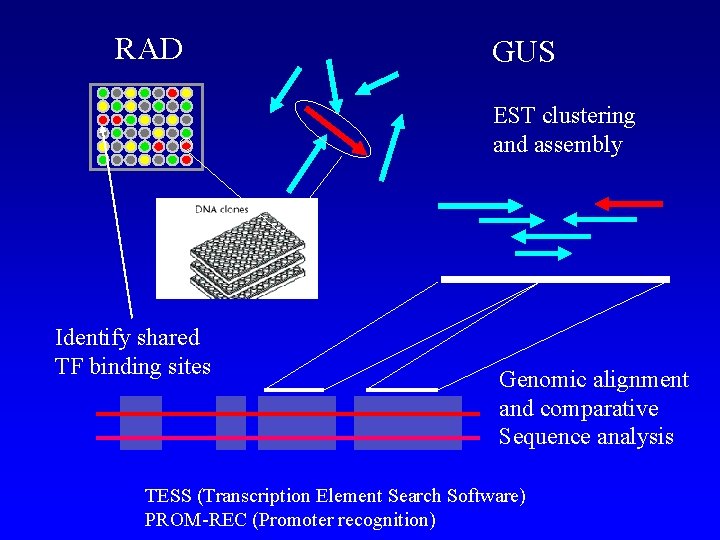
RAD GUS EST clustering and assembly Identify shared TF binding sites Genomic alignment and comparative Sequence analysis TESS (Transcription Element Search Software) PROM-REC (Promoter recognition)

Acknowledgements CBIL: Chris Overton Chris Stoeckert Vladimir Babenko Brian Brunk Jonathan Crabtree Sharon Diskin Greg Grant Yuri Kondrakhin Georgi Kostov Phil Le Elisabetta Manduchi Joan Mazzarelli Shannon Mc. Weeney Debbie Pinney Angel Pizarro Jonathan Schug Plasmo. DB collaborators: David Roos Martin Fraunholz Jesse Kissinger Jules Milgram Ross Koppel, Monash U. Malarial Genome Sequencing Consortium (Sanger Centre, Stanford U. , TIGR/NMRC) Allgenes. org collaborators: Ed Uberbacher, ORNL Doug Hyatt, ORNL EPCon. DB collaborators: Klaus Kaestner Marie Scearce Doug Melton, Harvard Alan Permutt, Wash. U Comparative Sequence Analysis Collaborators: Maja Bucan Shaying Zhao Whitehead/MIT Center for Genome Research

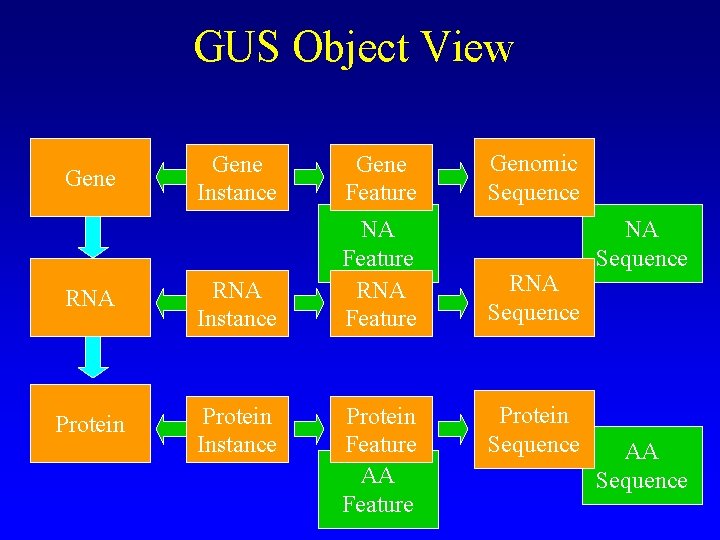
GUS Object View Gene Instance Gene Feature RNA Instance NA Feature RNA Feature Protein Instance Gene Protein Feature AA Feature Genomic Sequence RNA Sequence Protein Sequence NA Sequence AA Sequence

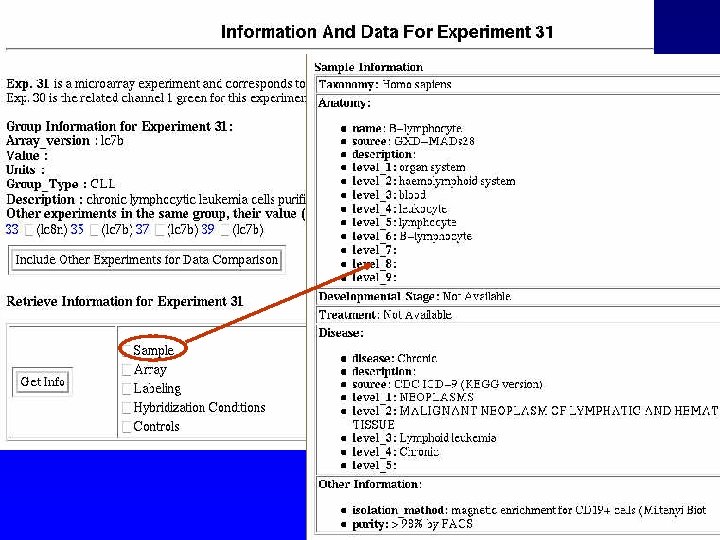
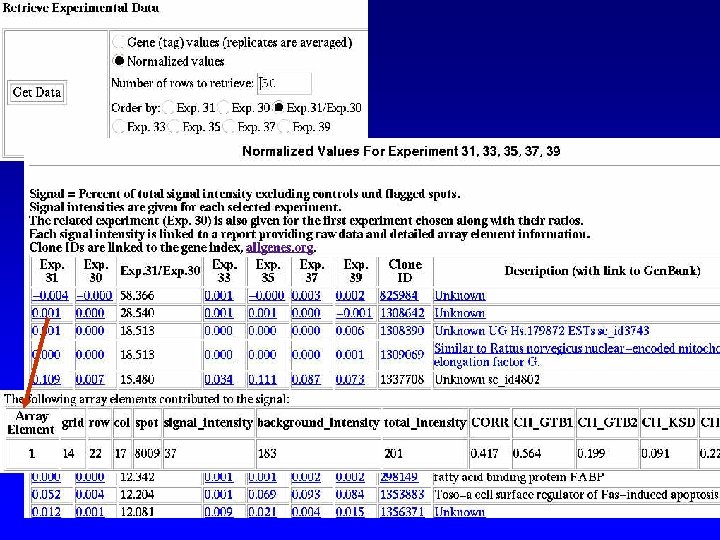
Query RAD by Sample or by Experiment
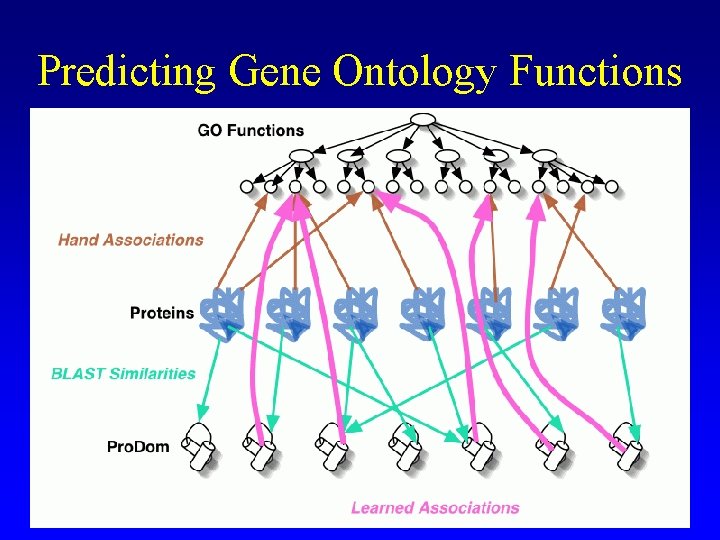
Predicting Gene Ontology Functions
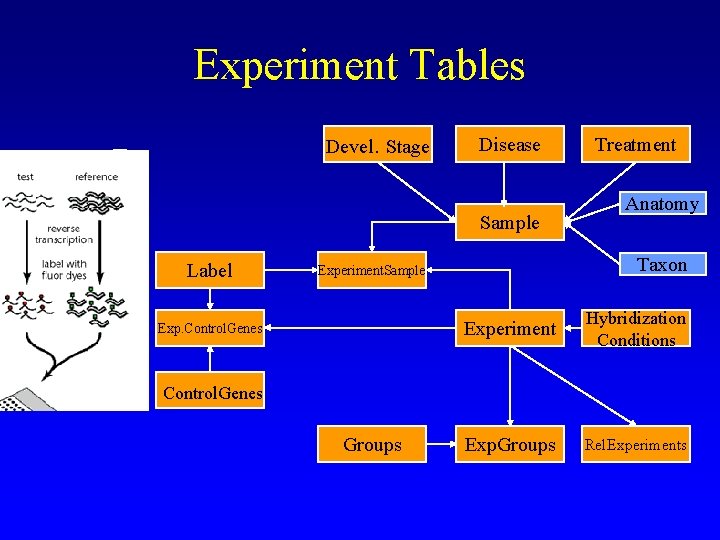
Experiment Tables Devel. Stage Disease Sample Label Anatomy Taxon Experiment. Sample Exp. Control. Genes Treatment Experiment Hybridization Conditions Exp. Groups Rel. Experiments Control. Genes Groups
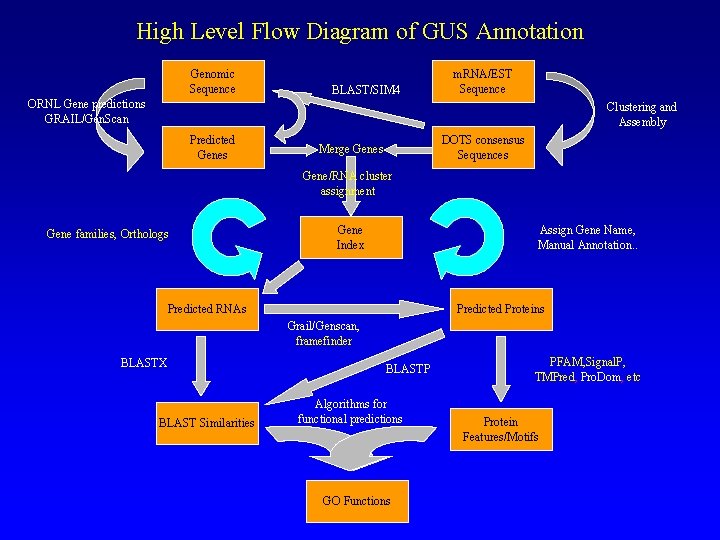
High Level Flow Diagram of GUS Annotation Genomic Sequence BLAST/SIM 4 m. RNA/EST Sequence ORNL Gene predictions GRAIL/Gen. Scan Clustering and Assembly Predicted Genes DOTS consensus Sequences Merge Genes Gene/RNA cluster assignment Gene families, Orthologs Gene Index Assign Gene Name, Manual Annotation. . Predicted RNAs Predicted Proteins Grail/Genscan, framefinder BLASTX BLAST Similarities BLASTP Algorithms for functional predictions GO Functions PFAM, Signal. P, TMPred, Pro. Dom, etc Protein Features/Motifs
- Slides: 26