Lecture Power Point to accompany Molecular Biology Fourth
Lecture Power. Point to accompany Molecular Biology Fourth Edition Robert F. Weaver Chapter 12 Transcription Activators in Eukaryotes Copyright © The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display.
12. 1 Categories of Activators • Activators can stimulate or inhibit transcription by RNA polymerase II • Structure is composed of at least 2 functional domains – DNA-binding domain – Transcription-activation domain – Many also have a dimerization domain 2
DNA-Binding Domains • Protein domain is an independently folded region of a protein • DNA-binding domains have DNA-binding motif – Part of the domain having characteristic shape specialized for specific DNA binding – Most DNA-binding motifs fall into 3 classes 3
Zinc-Containing Modules • There at least 3 kinds of zinccontaining modules that act as DNAbinding motifs • All use one or more zinc ions to create a shape to fit an a-helix of the motif into the DNA major groove – Zinc fingers – Zinc modules – Modules containing 2 zinc and 6 cysteines 4
Homeodomains • These domains contain about 60 amino acids • Resemble the helix-turn-helix proteins in structure and function • Found in a variety of activators • Originally identified in homeobox proteins regulating fruit fly development 5
b. ZIP and b. HLH Motifs • A number of transcription factors have a highly basic DNA-binding motif linked to protein dimerization motifs – Leucine zippers – Helix-loop-helix • Examples include: – CCAAT/enhancer-binding protein – Myo. D protein 6
Transcription-Activating Domains • Most activators have one of these domains • Some have more than one – Acidic domains such as yeast GAL 4 with 11 acidic amino acids out of 49 amino acids in the domain – Glutamine-rich domains include Sp 1 having 2 that are 25% glutamine – Proline-rich domains such as CTF which has a domain of 84 amino acids, 19 proline 7
12. 2 Structures of the DNABinding Motifs of Activators • DNA-binding domains have well-defined structures • X-ray crystallographic studies have shown how these structures interact with their DNA targets • Interaction domains forming dimers, or tetramers, have also been described • Most classes of DNA-binding proteins can’t bind DNA in monomer form 8

Zinc Fingers • Described by Klug in TFIIIA • Nine repeats of a 30 -residue element: – 2 closely spaced cysteines followed 12 amino acids later by 2 closely spaced histidines – Coordination of amino acids to the metal helps form the finger-shaped structure – Rich in zinc, enough for 1 zinc ion per repeat – Specific recognition between the zinc finger and its DNA target occurs in the major groove 9
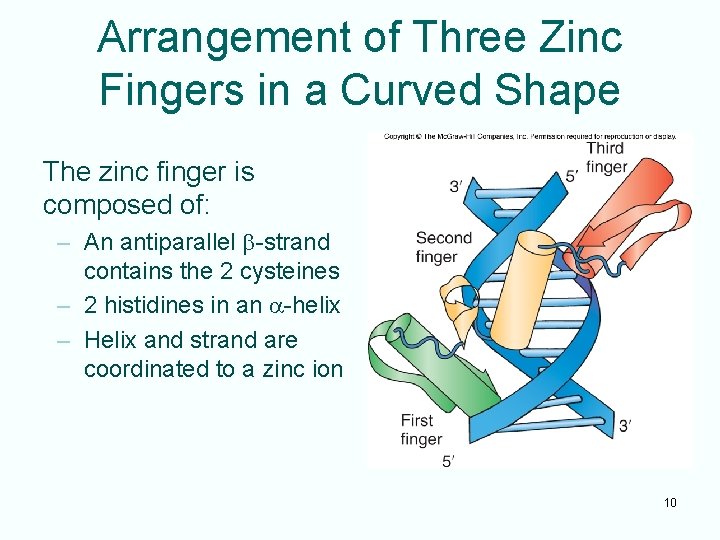
Arrangement of Three Zinc Fingers in a Curved Shape The zinc finger is composed of: – An antiparallel b-strand contains the 2 cysteines – 2 histidines in an a-helix – Helix and strand are coordinated to a zinc ion 10
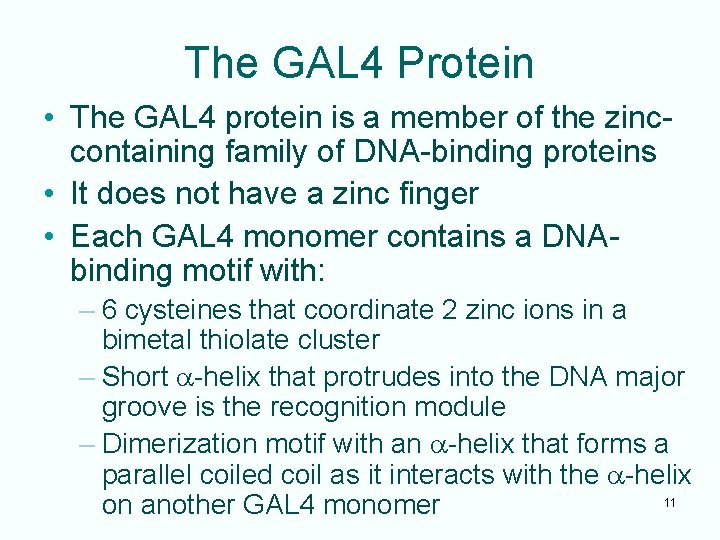
The GAL 4 Protein • The GAL 4 protein is a member of the zinccontaining family of DNA-binding proteins • It does not have a zinc finger • Each GAL 4 monomer contains a DNAbinding motif with: – 6 cysteines that coordinate 2 zinc ions in a bimetal thiolate cluster – Short a-helix that protrudes into the DNA major groove is the recognition module – Dimerization motif with an a-helix that forms a parallel coiled coil as it interacts with the a-helix 11 on another GAL 4 monomer
The Nuclear Receptors • A third class of zinc module is the nuclear receptor • This type of protein interacts with a variety of endocrine-signaling molecules • Protein plus endocrine molecule forms a complex that functions as an activator by binding to hormone response elements and stimulating transcription of associated genes 12
Type I Nuclear Receptors • These receptors reside in the cytoplasm bound to another protein • When receptors bind to their hormone ligands: – Release their cytoplasmic protein partners – Move to nucleus – Bind to enhancers – Act as activators 13
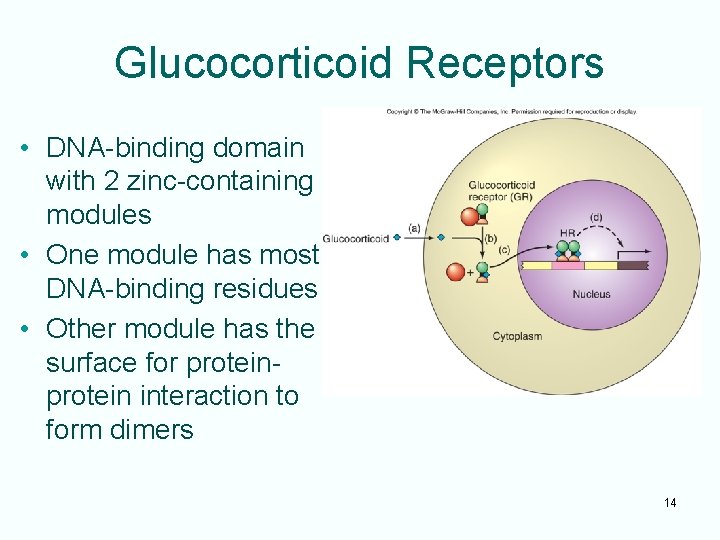
Glucocorticoid Receptors • DNA-binding domain with 2 zinc-containing modules • One module has most DNA-binding residues • Other module has the surface for protein interaction to form dimers 14

Types II and III Nuclear Receptors • Type II nuclear receptors stay within the nucleus • Bound to target DNA sites • Without ligands the receptors repress gene activity • When receptors bind ligands, they activate transcription • Type III receptors are “orphan” whose ligands are not yet identified 15
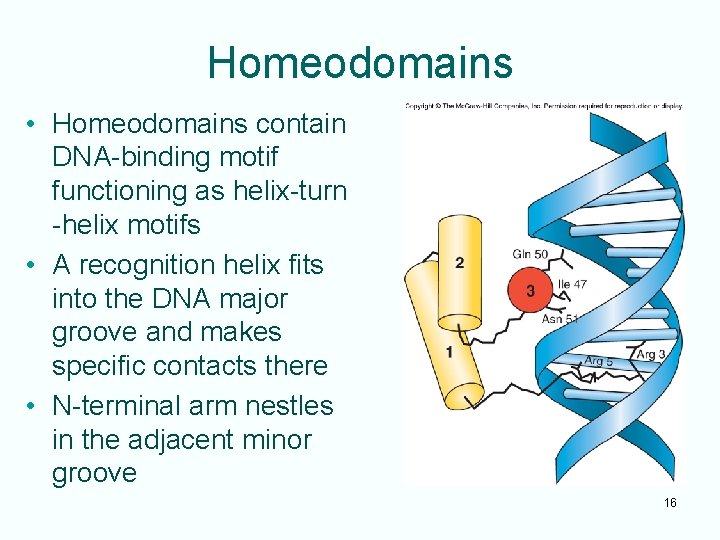
Homeodomains • Homeodomains contain DNA-binding motif functioning as helix-turn -helix motifs • A recognition helix fits into the DNA major groove and makes specific contacts there • N-terminal arm nestles in the adjacent minor groove 16
The b. ZIP and b. HLH Domains • b. ZIP proteins dimerize through a leucine zipper – This puts the adjacent basic regions of each monomer in position to embrace DNA target like a pair of tongs • b. HLH proteins dimerize through a helix-loophelix motif – Allows basic parts of each long helix to grasp the DNA target site • b. HLH and b. HLH-ZIP domains bind to DNA in the same way, later have extra dimerization potential due to their leucine zippers 17
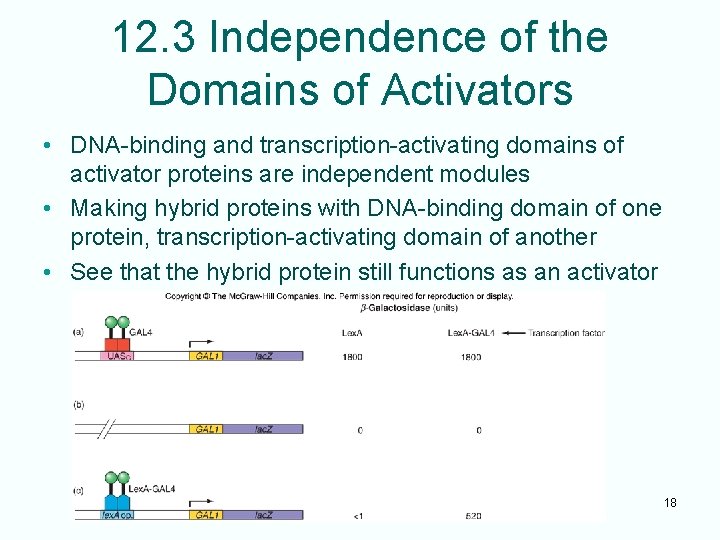
12. 3 Independence of the Domains of Activators • DNA-binding and transcription-activating domains of activator proteins are independent modules • Making hybrid proteins with DNA-binding domain of one protein, transcription-activating domain of another • See that the hybrid protein still functions as an activator 18
12. 4 Functions of Activators • Bacterial core RNA polymerase is incapable of initiating meaningful transcription • RNA polymerase holoenzyme can catalyze basal level transcription – Often insufficient at weak promoters – Cells have activators to boost basal transcription to higher level in a process called recruitment 19

Eukaryotic Activators • Eukaryotic activators also recruit RNA polymerase to promoters • Stimulate binding of general transcription factors and RNA polymerase to a promoter • 2 hypotheses for recruitment: – General TF cause a stepwise build-up of preinitiation complex – General TF and other proteins are already bound to polymerase in a complex called RNA polymerase holoenzyme 20
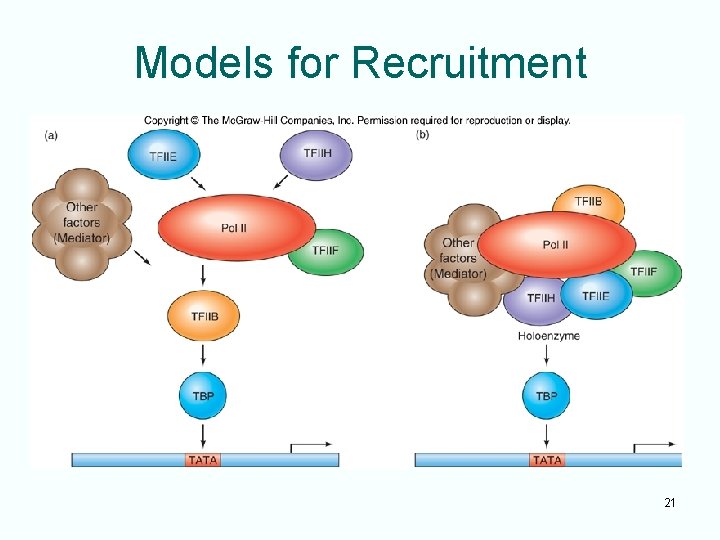
Models for Recruitment 21
Recruitment of TFIID • Acidic transcription-activating domain of the herpes virus transcription factor VP 16 binds to TFIID under affinity chromatography conditions • TFIID is rate-limiting for transcription in some systems • TFIID is the important target of the VP 16 transcription-activating domain 22

Recruitment of the Holoenzyme • Activation in some yeast promoters appears to function by recruitment of holoenzyme • This is an alternative to the recruitment of individual components of the holoenzyme one at a time • Some evidence suggests that recruitment of the holoenzyme as a unit is not common 23
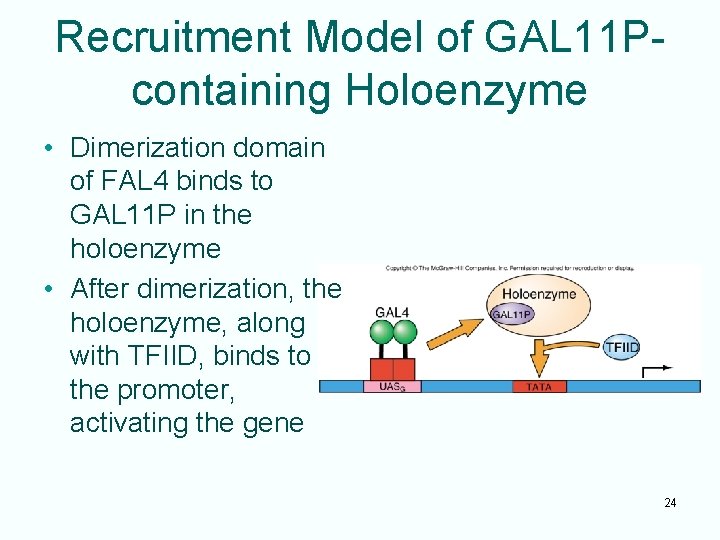
Recruitment Model of GAL 11 Pcontaining Holoenzyme • Dimerization domain of FAL 4 binds to GAL 11 P in the holoenzyme • After dimerization, the holoenzyme, along with TFIID, binds to the promoter, activating the gene 24
12. 5 Interaction Among Activators • General transcription factors must interact to form the preinitiation complex • Activators and general transcription factors also interact • Activators usually interact with one another in activating a gene – Individual factors interact to form a protein dimer facilitating binding to a single DNA target site – Specific factors bound to different DNA target 25 sites can collaborate in activating a gene
Dimerization • Dimerization is a great advantage to an activator • Dimerization increases the affinity between activator and its DNA target • Some activators form homodimers • Heterodimers are also formed – Products of the jun and fos genes form a heterodimer 26

Action at a Distance • Bacterial and eukaryotic enhancers stimulate transcription even though located some distance from their promoters • Four hypotheses attempt to explain the ability of enhancers to act at a distance – Change in topology – Sliding – Looping – Facilitated tracking 27
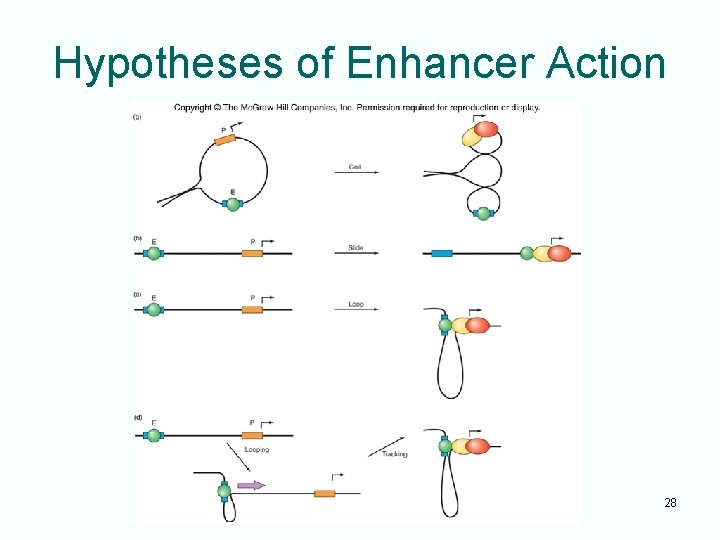
Hypotheses of Enhancer Action 28

Complex Enhancers • Many genes can have more than one activator-binding site permitting them to respond to multiple stimuli • Each of the activators that bind at these sites must be able to interact with the preinitiation complex assembling at the promoter, likely by looping out any intervening DNA 29
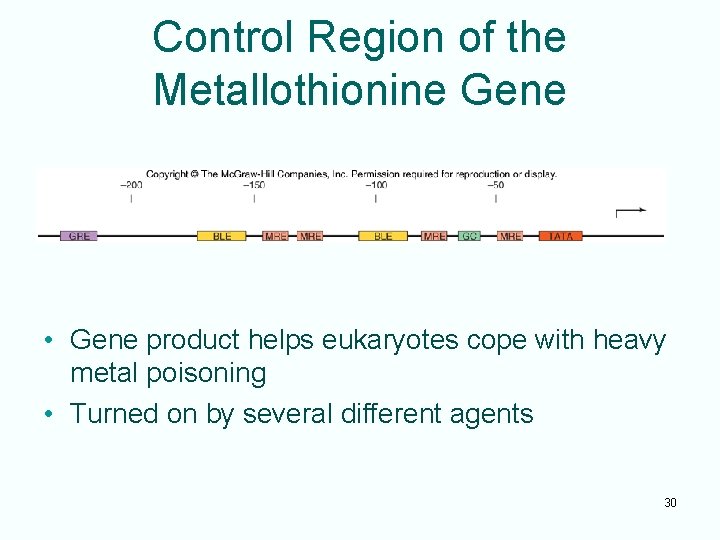
Control Region of the Metallothionine Gene • Gene product helps eukaryotes cope with heavy metal poisoning • Turned on by several different agents 30
Architectural Transcription Factors Architectural transcription factors are those transcription factors whose sole or main purpose seems to be to change the shape of a DNA control region so that other proteins can interact successfully to stimulate transcription 31
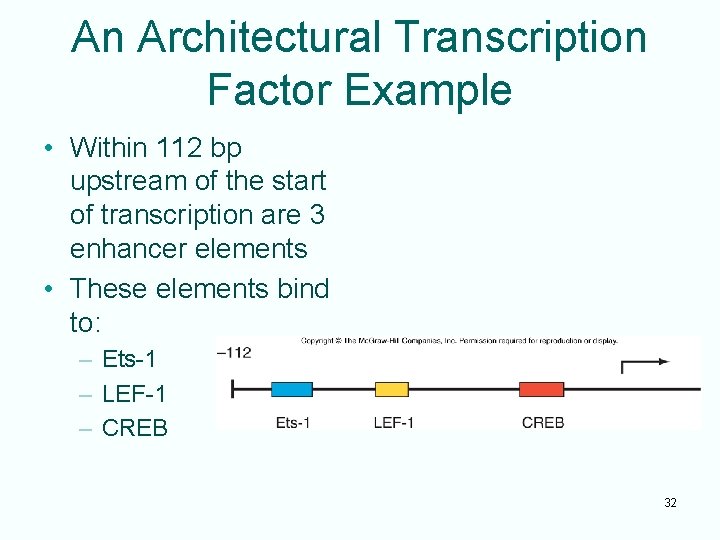
An Architectural Transcription Factor Example • Within 112 bp upstream of the start of transcription are 3 enhancer elements • These elements bind to: – Ets-1 – LEF-1 – CREB 32
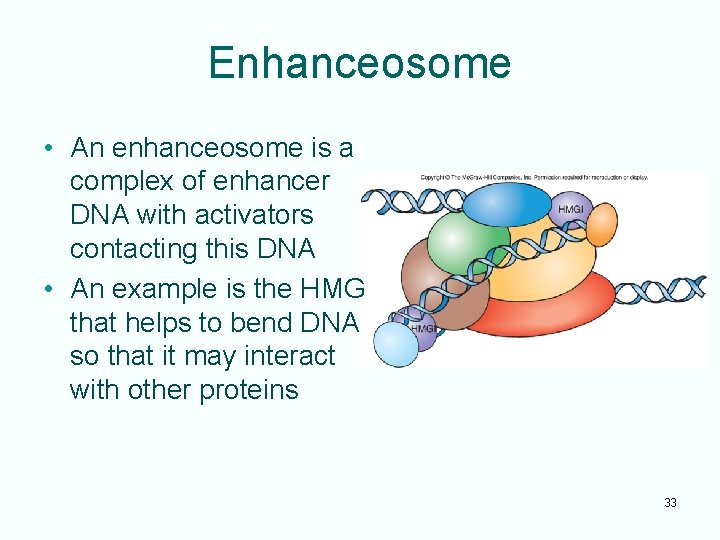
Enhanceosome • An enhanceosome is a complex of enhancer DNA with activators contacting this DNA • An example is the HMG that helps to bend DNA so that it may interact with other proteins 33
DNA Bending Aids Protein Binding • The activator LEF-1 binds to the minor groove of its DNA target through its HMG domain and induces strong bending of DNA • LEF-1 does not enhance transcription by itself • Bending it induces helps other activators bind and interact with activators and general transcription factors 34
Examples of Architectural Transcription Factors • Besides LEF-1, HMG I(Y) plays a similar role in the human interferon-b control gene • For the IFN-b enhancer, activation seems to require cooperative binding of several activators, including HMG I(Y) to form an enhanceosome with a specific shape 35
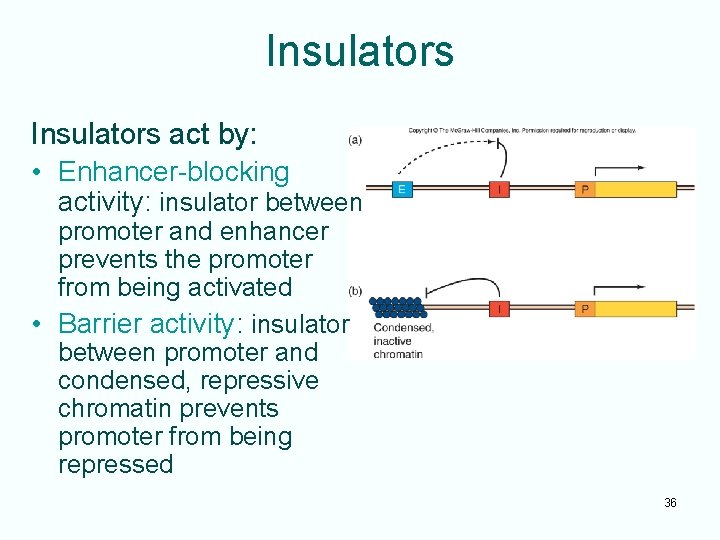
Insulators act by: • Enhancer-blocking activity: insulator between promoter and enhancer prevents the promoter from being activated • Barrier activity: insulator between promoter and condensed, repressive chromatin prevents promoter from being repressed 36
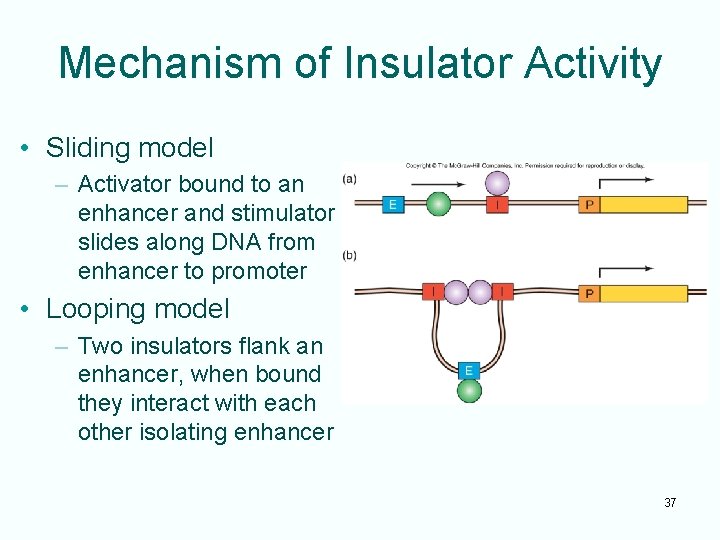
Mechanism of Insulator Activity • Sliding model – Activator bound to an enhancer and stimulator slides along DNA from enhancer to promoter • Looping model – Two insulators flank an enhancer, when bound they interact with each other isolating enhancer 37
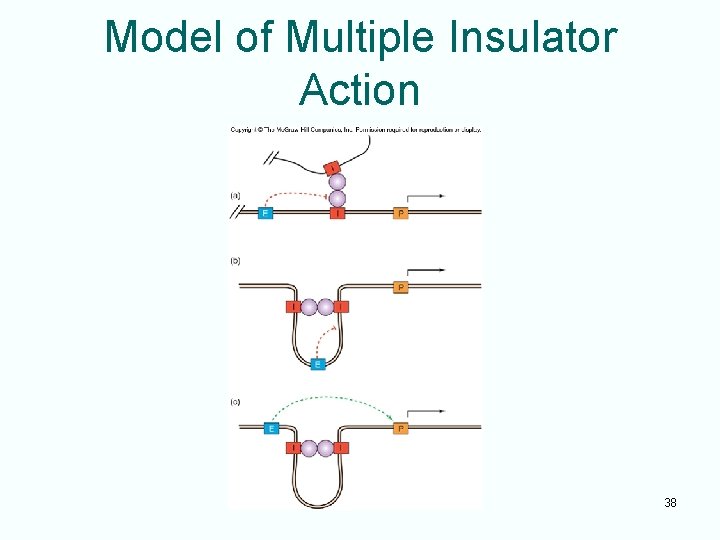
Model of Multiple Insulator Action 38
12. 6 Regulation of Transcription Factors • Phosphorylation of activators can allow them to interact with coactivators that in turn stimulate transcription • Ubiquitylation of transcription factors can mark them for – Destruction by proteolysis – Stimulation of activity • Sumoylation is the attachment of the polypeptide SUMO which can target for incorporation into compartments of the nucleus • Methylation and acetylation can modulate activity 39
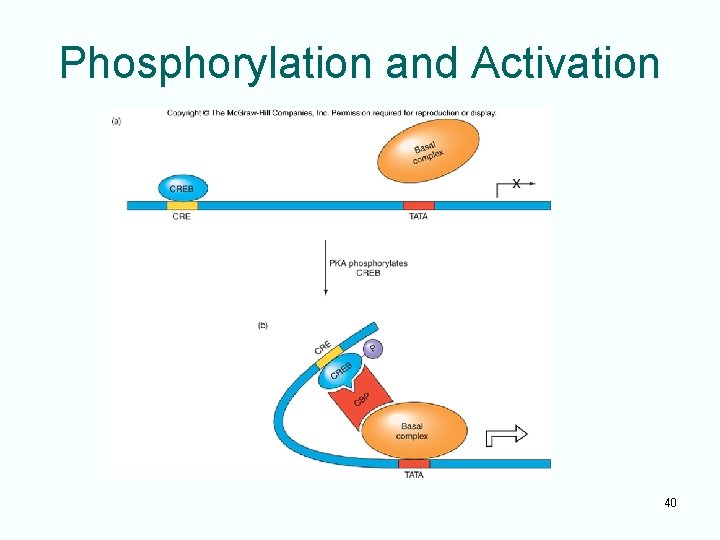
Phosphorylation and Activation Replace this area with Figure 12. 33: A model for activation of a CRElinked gene 40
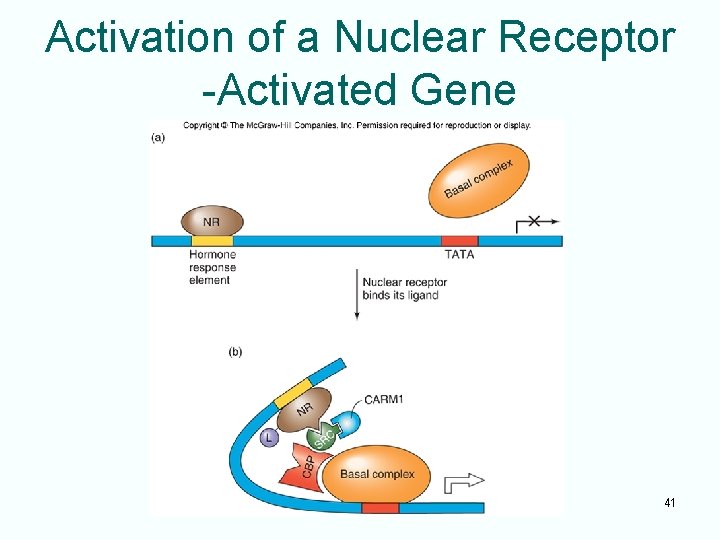
Activation of a Nuclear Receptor -Activated Gene 41

Ubiquitylation • Ubiquitylation, especially monoubiquitylation, of some activators can have an activating effect • Polyubiquitylation marks these same proteins for destruction • Proteins from the 19 S regulatory particle of the proteasome can stimulate transcription 42
Activator Sumoylation • Sumoylation is the addition of one or more copies of the 101 -amino acid polypeptide SUMO (Small Ubiquitin-Related Modifier) to lysine residues on a protein • Process is similar to ubiquitylation • Results quite different – sumoylated activators are targeted to a specific nuclear compartment that keeps them stable 43
Activator Acetylation • Nonhistone activators and repressors can be acetylated by HATs • HAT is the enzyme histone acetyltransferase which can act on nonhistone activators and repressors • Such acetylation can have either positive or negative effects 44
Signal Transduction Pathways • Signal transduction pathways begin with a signaling molecule interacting with a receptor on the cell surface • This interaction sends the signal into the cell and frequently leads to altered gene expression • Many signal transduction pathways rely on protein phosphorylation to pass the signal from one protein to another • This leads to signal amplification at each step 45
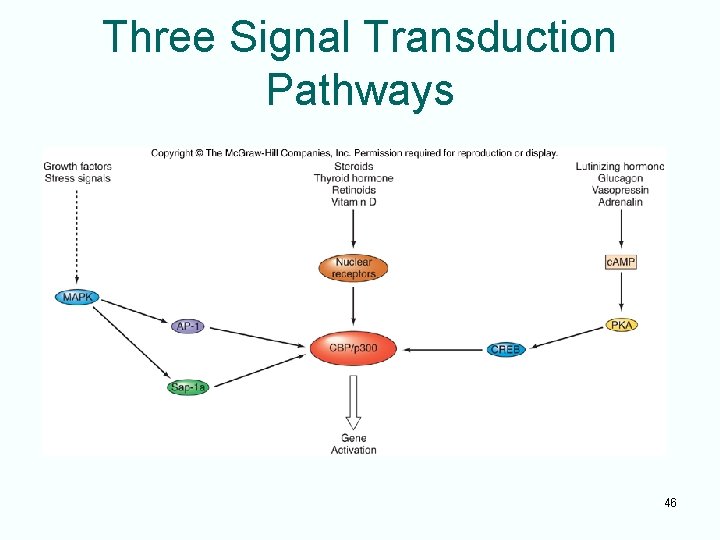
Three Signal Transduction Pathways 46
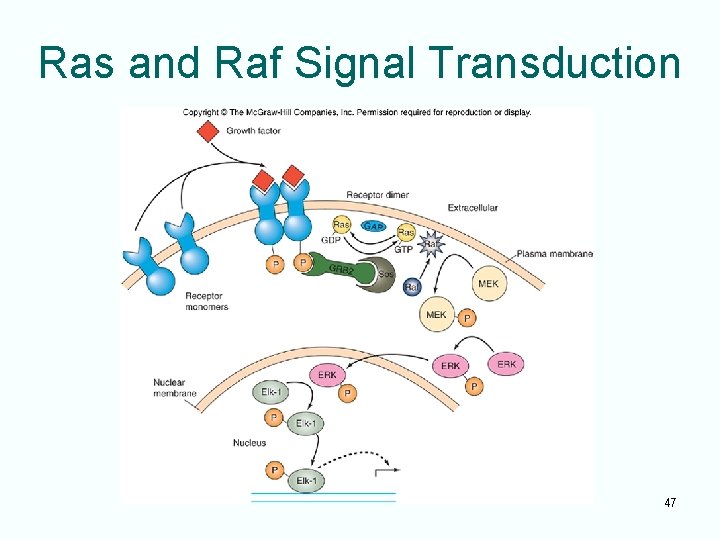
Ras and Raf Signal Transduction 47
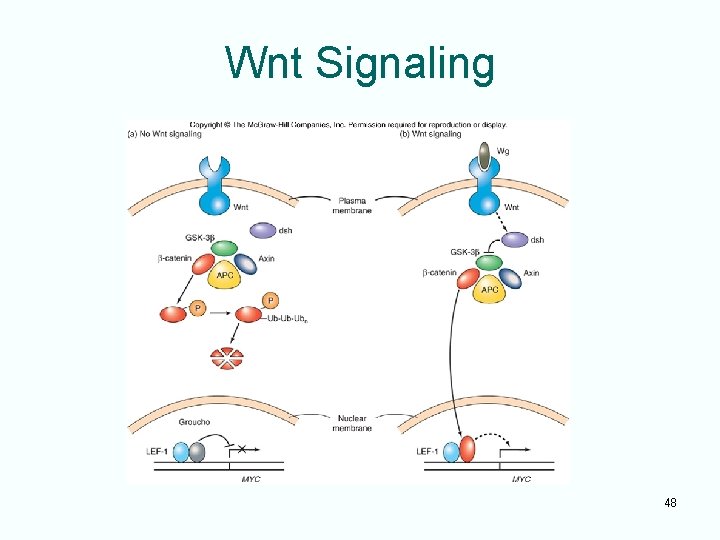
Wnt Signaling 48
- Slides: 48