Lecture Data Mining in R 732 A 44



![Neural network • Input nodes, input layer • [Hidden nodes, Hidden layer(s)] • Output Neural network • Input nodes, input layer • [Hidden nodes, Hidden layer(s)] • Output](https://slidetodoc.com/presentation_image_h/014e29960f0bbb10c6a67ff0a0534a5a/image-21.jpg)
![K-means • One way to visualize plot(m 5$linoleic, m 5$eicosenoic, col=res$cluster); points(res$centers[, 1], res$centers[, K-means • One way to visualize plot(m 5$linoleic, m 5$eicosenoic, col=res$cluster); points(res$centers[, 1], res$centers[,](https://slidetodoc.com/presentation_image_h/014e29960f0bbb10c6a67ff0a0534a5a/image-27.jpg)


- Slides: 33

Lecture Data Mining in R 732 A 44 Programming in R

Logistic regression: two classes • Consider Logistic model with one predictor X=Price of the car Y=Equipment • Logistic model • Use function glm(formula, family, data) – Formula: Response~Model • Model consists of a+b (addition), a: b (interaction terms, a*b (addition and interaction). All predictors – Family: specify binomial 732 A 44 Programming in R

Logistic regression: two classes reg<-glm(X 3. . . Equipment~Price. in. SEK. , family=binomial, data=mydata); 732 A 44 Programming in R

Logistic regression: several predictors Data about contraceptive use – Several analysis plots can be obtained by plot(lrfit) – Response: matrix success/failure 732 A 44 Programming in R

Logistic regression Further comments • Nominal logistic regressions (library mlogit, function mlogit) • Stepwise model selection: step() function. • Prediction: predict() function 732 A 44 Programming in R

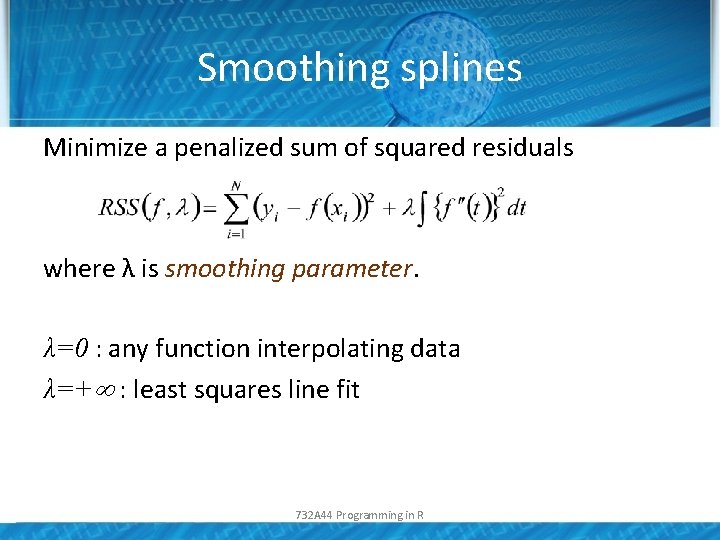
Smoothing splines Minimize a penalized sum of squared residuals where λ is smoothing parameter. λ=0 : any function interpolating data λ=+ : least squares line fit 732 A 44 Programming in R

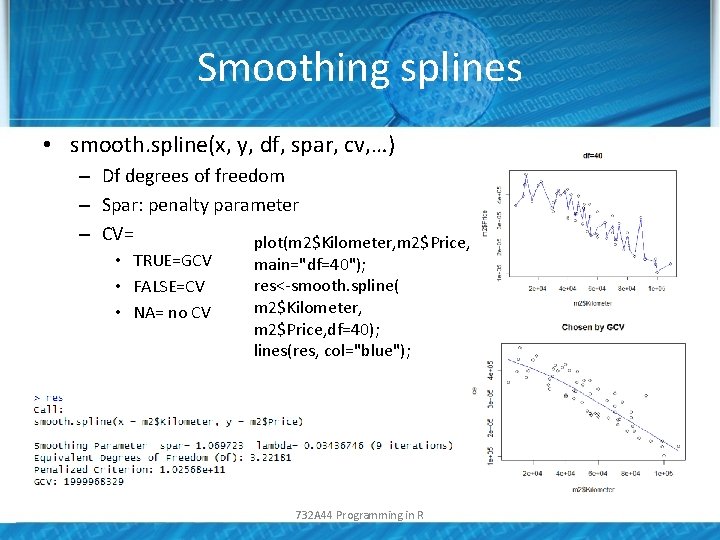
Smoothing splines • smooth. spline(x, y, df, spar, cv, …) – Df degrees of freedom – Spar: penalty parameter – CV= plot(m 2$Kilometer, m 2$Price, • TRUE=GCV • FALSE=CV • NA= no CV main="df=40"); res<-smooth. spline( m 2$Kilometer, m 2$Price, df=40); lines(res, col="blue"); 732 A 44 Programming in R

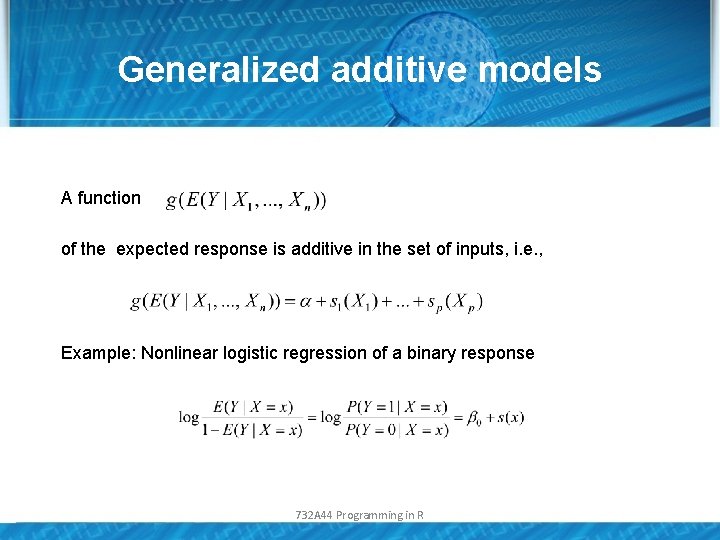
Generalized additive models A function of the expected response is additive in the set of inputs, i. e. , Example: Nonlinear logistic regression of a binary response 732 A 44 Programming in R

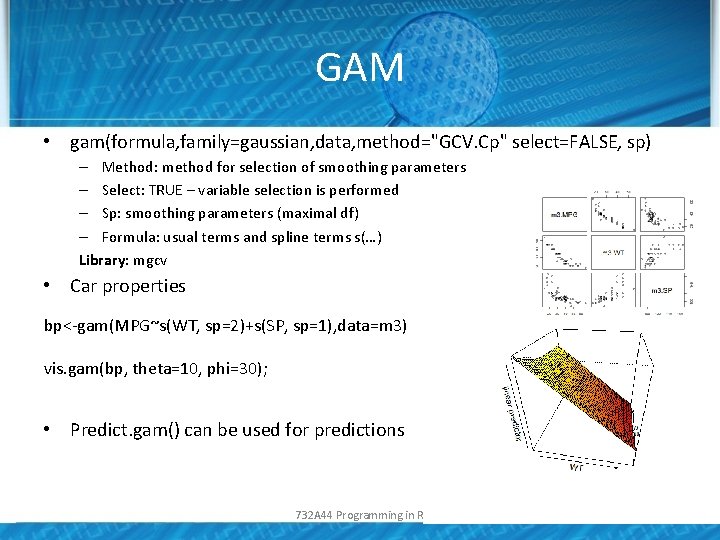
GAM • gam(formula, family=gaussian, data, method="GCV. Cp" select=FALSE, sp) – Method: method for selection of smoothing parameters – Select: TRUE – variable selection is performed – Sp: smoothing parameters (maximal df) – Formula: usual terms and spline terms s(…) Library: mgcv • Car properties bp<-gam(MPG~s(WT, sp=2)+s(SP, sp=1), data=m 3) vis. gam(bp, theta=10, phi=30); • Predict. gam() can be used for predictions 732 A 44 Programming in R

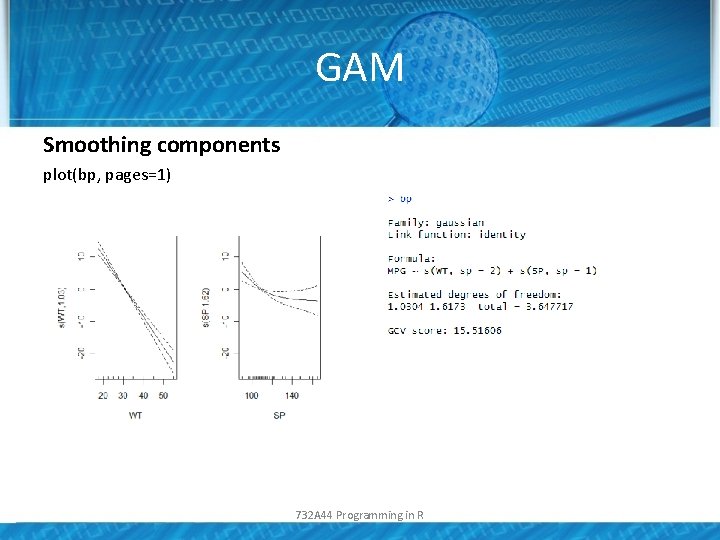
GAM Smoothing components plot(bp, pages=1) 732 A 44 Programming in R

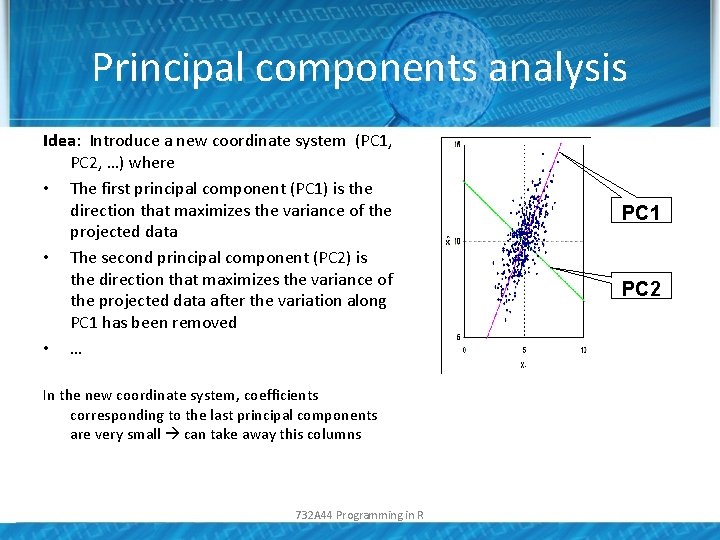
Principal components analysis Idea: Introduce a new coordinate system (PC 1, PC 2, …) where • The first principal component (PC 1) is the direction that maximizes the variance of the projected data • The second principal component (PC 2) is the direction that maximizes the variance of the projected data after the variation along PC 1 has been removed • … In the new coordinate system, coefficients corresponding to the last principal components are very small can take away this columns 732 A 44 Programming in R PC 1 PC 2

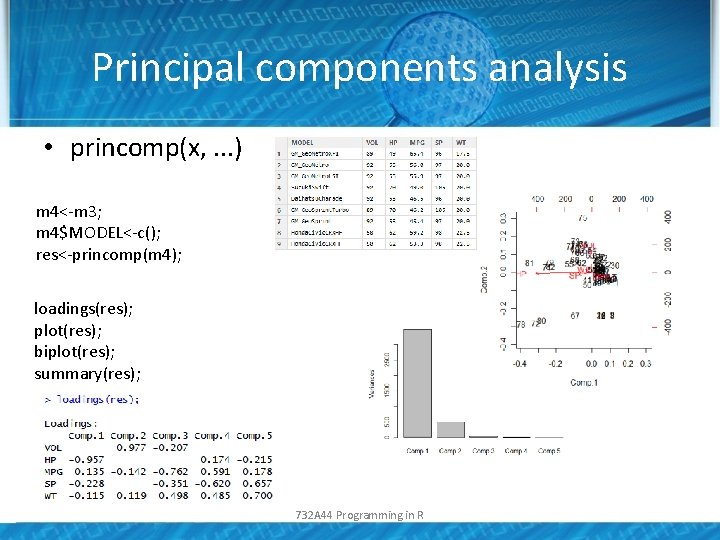
Principal components analysis • princomp(x, . . . ) m 4<-m 3; m 4$MODEL<-c(); res<-princomp(m 4); loadings(res); plot(res); biplot(res); summary(res); 732 A 44 Programming in R

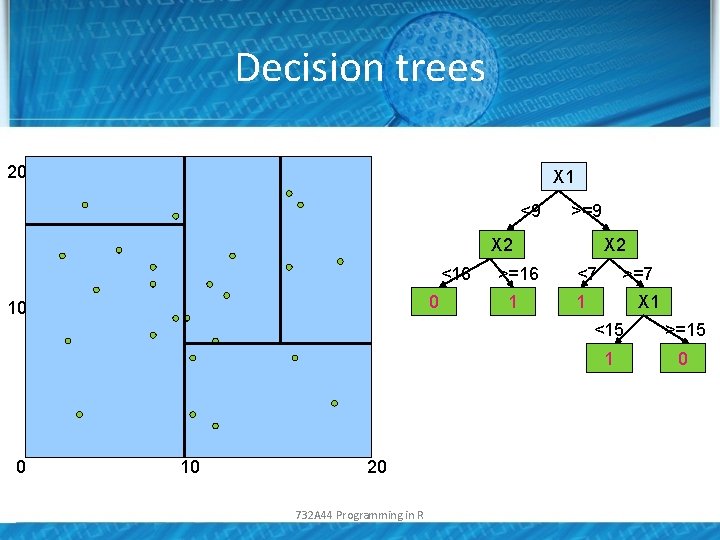
Decision trees 20 X 1 <9 >=9 X 2 <16 0 10 20 732 A 44 Programming in R >=16 1 X 2 <7 >=7 1 X 1 <15 >=15 1 0

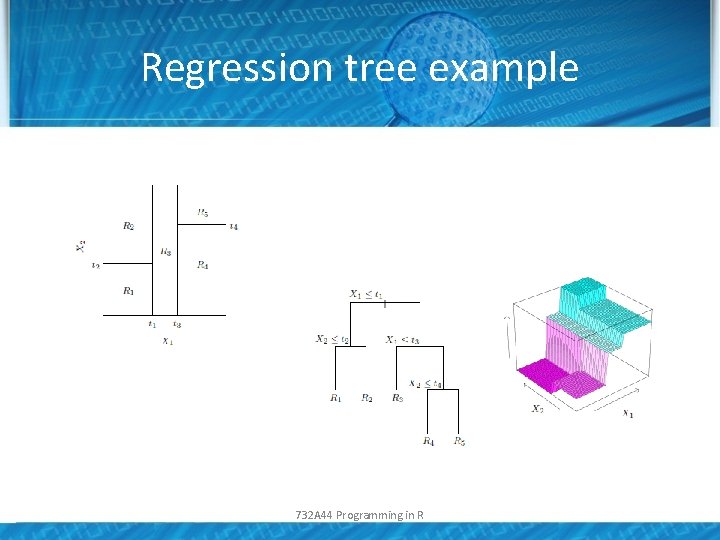
Regression tree example 732 A 44 Programming in R

Training-validation-test • Training-validation (60/40) sub <- sample(nrow(m 2), floor(nrow(m 2) * 0. 6)) training <- m 2[sub, ] validation <- m 2[-sub, ] • If training-validation-test is required, use similar strategy 732 A 44 Programming in R

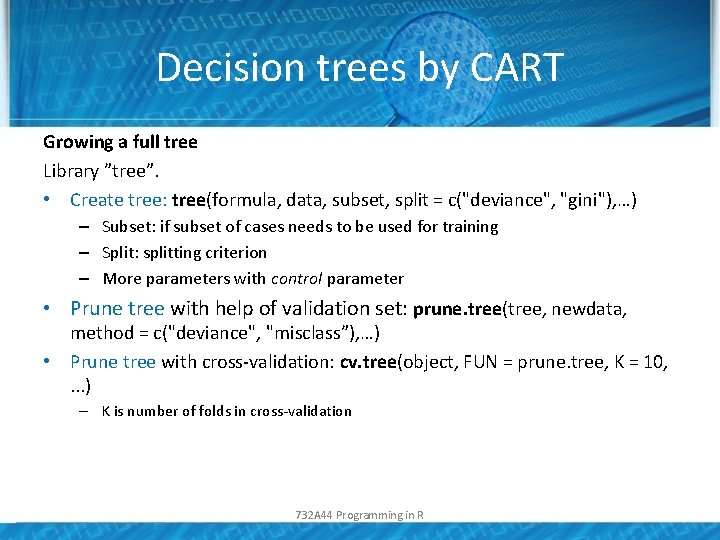
Decision trees by CART Growing a full tree Library ”tree”. • Create tree: tree(formula, data, subset, split = c("deviance", "gini"), …) – Subset: if subset of cases needs to be used for training – Split: splitting criterion – More parameters with control parameter • Prune tree with help of validation set: prune. tree(tree, newdata, method = c("deviance", "misclass”), …) • Prune tree with cross-validation: cv. tree(object, FUN = prune. tree, K = 10, . . . ) – K is number of folds in cross-validation 732 A 44 Programming in R

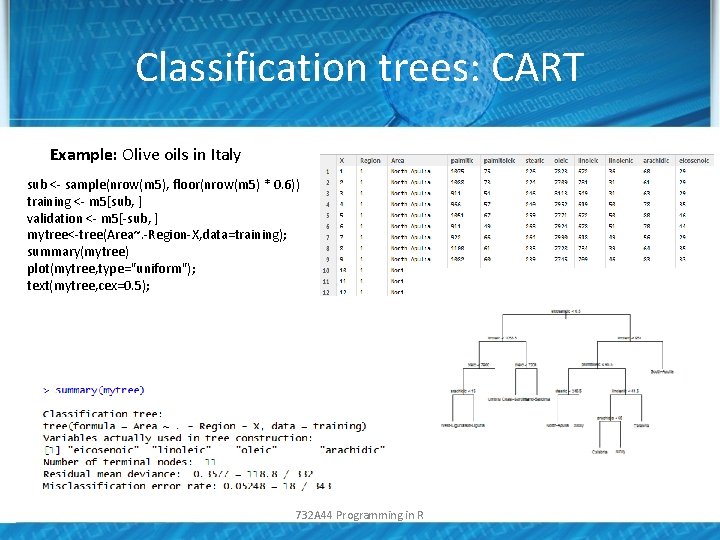
Classification trees: CART Example: Olive oils in Italy sub <- sample(nrow(m 5), floor(nrow(m 5) * 0. 6)) training <- m 5[sub, ] validation <- m 5[-sub, ] mytree<-tree(Area~. -Region-X, data=training); summary(mytree) plot(mytree, type="uniform"); text(mytree, cex=0. 5); 732 A 44 Programming in R

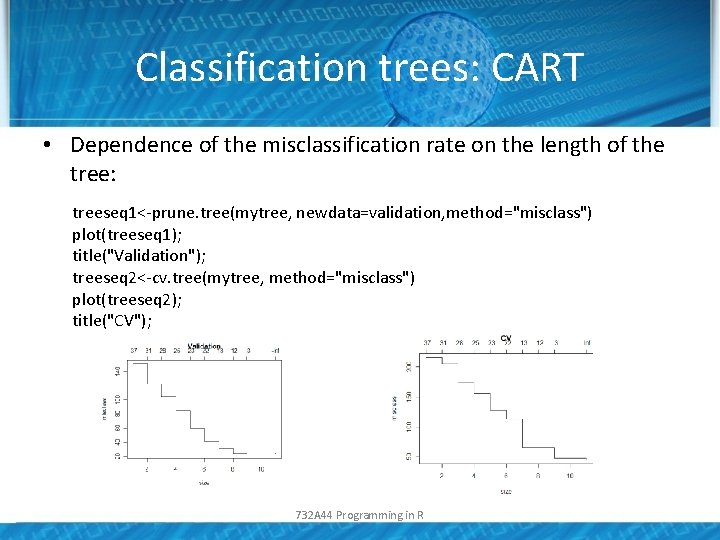
Classification trees: CART • Dependence of the misclassification rate on the length of the tree: treeseq 1<-prune. tree(mytree, newdata=validation, method="misclass") plot(treeseq 1); title("Validation"); treeseq 2<-cv. tree(mytree, method="misclass") plot(treeseq 2); title("CV"); 732 A 44 Programming in R

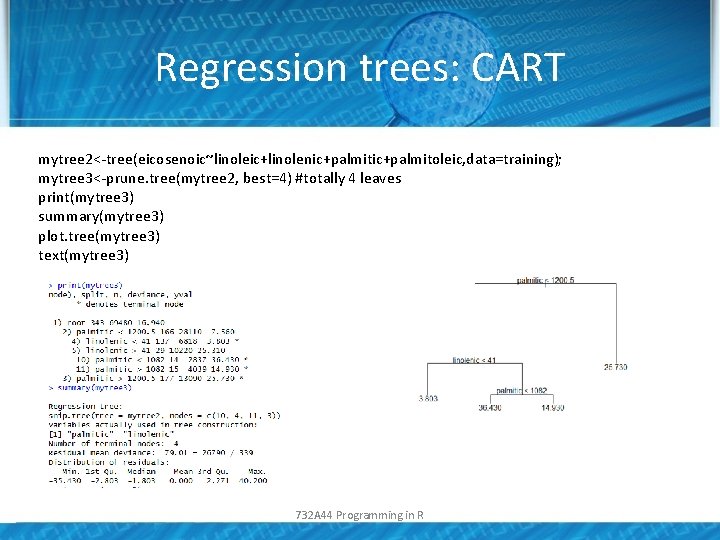
Regression trees: CART mytree 2<-tree(eicosenoic~linoleic+linolenic+palmitoleic, data=training); mytree 3<-prune. tree(mytree 2, best=4) #totally 4 leaves print(mytree 3) summary(mytree 3) plot. tree(mytree 3) text(mytree 3) 732 A 44 Programming in R

Decision trees: other techniques • Conditional inference trees Library: party training$X<-c(); training$Area<-c(); mytree 4<-ctree(Region~. , data=training); print(mytree 4) plot(mytree 4, type= "simple"); # gives nice plots • CART, another library ”rpart” 732 A 44 Programming in R
![Neural network Input nodes input layer Hidden nodes Hidden layers Output Neural network • Input nodes, input layer • [Hidden nodes, Hidden layer(s)] • Output](https://slidetodoc.com/presentation_image_h/014e29960f0bbb10c6a67ff0a0534a5a/image-21.jpg)
Neural network • Input nodes, input layer • [Hidden nodes, Hidden layer(s)] • Output nodes, output layer • Weights • Activation functions • Combination functions z 1 x 1 732 A 44 Programming in R … f 1 z 2 x 2 f. K … … z. M xp

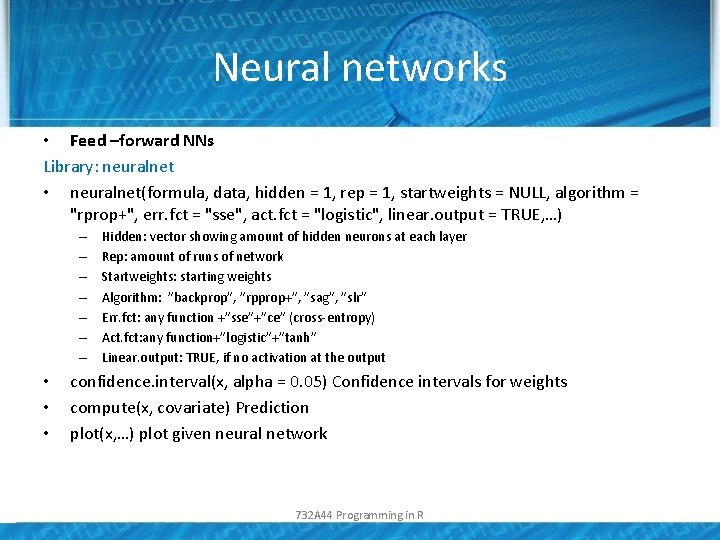
Neural networks • Feed –forward NNs Library: neuralnet • neuralnet(formula, data, hidden = 1, rep = 1, startweights = NULL, algorithm = "rprop+", err. fct = "sse", act. fct = "logistic", linear. output = TRUE, …) – – – – • • • Hidden: vector showing amount of hidden neurons at each layer Rep: amount of runs of network Startweights: starting weights Algorithm: ”backprop”, ”rpprop+”, ”sag”, ”slr” Err. fct: any function +”sse”+”ce” (cross-entropy) Act. fct: any function+”logistic”+”tanh” Linear. output: TRUE, if no activation at the output confidence. interval(x, alpha = 0. 05) Confidence intervals for weights compute(x, covariate) Prediction plot(x, …) plot given neural network 732 A 44 Programming in R

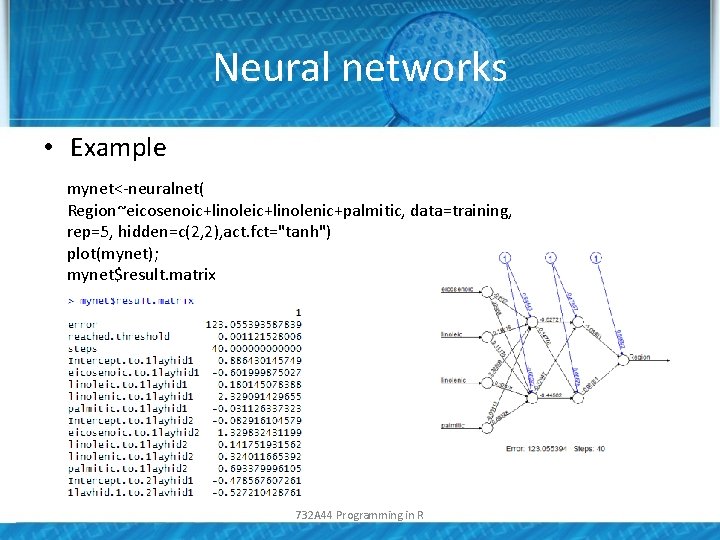
Neural networks • Example mynet<-neuralnet( Region~eicosenoic+linolenic+palmitic, data=training, rep=5, hidden=c(2, 2), act. fct="tanh") plot(mynet); mynet$result. matrix 732 A 44 Programming in R

Neural networks • Prediction with compute() • Finding misclassification rate: table(true_values, predicted values) – not only for neural networks • Another package, ready for qualitative response (classical nnet): mynet 1<-nnet( Region~eicosenoic+linoleic, data=training, size=3); coef(mynet 1) predict(mynet 1, data=validation); 732 A 44 Programming in R

Clustering • Purpose is to identify groups of observations into intput space (separated) – K-means – Hierarchical – Density-based 732 A 44 Programming in R

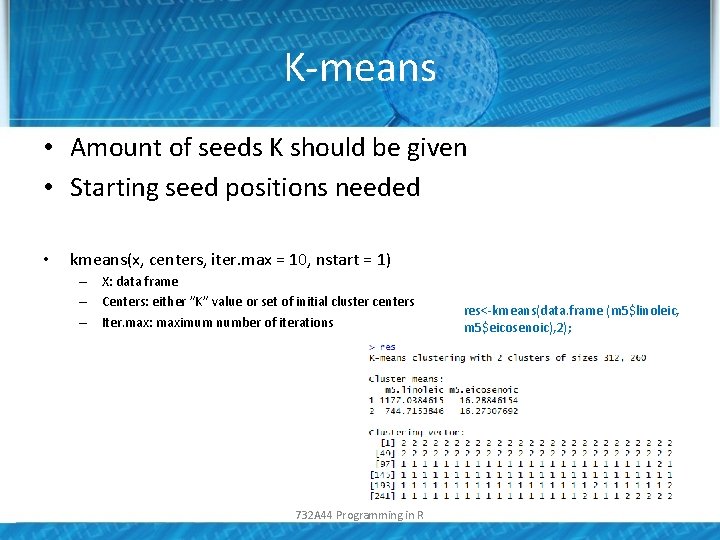
K-means • Amount of seeds K should be given • Starting seed positions needed • kmeans(x, centers, iter. max = 10, nstart = 1) – X: data frame – Centers: either ”K” value or set of initial cluster centers – Iter. max: maximum number of iterations 732 A 44 Programming in R res<-kmeans(data. frame (m 5$linoleic, m 5$eicosenoic), 2);
![Kmeans One way to visualize plotm 5linoleic m 5eicosenoic colrescluster pointsrescenters 1 rescenters K-means • One way to visualize plot(m 5$linoleic, m 5$eicosenoic, col=res$cluster); points(res$centers[, 1], res$centers[,](https://slidetodoc.com/presentation_image_h/014e29960f0bbb10c6a67ff0a0534a5a/image-27.jpg)
K-means • One way to visualize plot(m 5$linoleic, m 5$eicosenoic, col=res$cluster); points(res$centers[, 1], res$centers[, 2], col = 1: 2, pch = 8, cex=2) 732 A 44 Programming in R

Hierarchical clustering • Agglomerative – Place each point into a single cluster – Merge nearest clusters until you get 1 cluster • Meaning of ”two objects are close”? – Measure of proximity (ex: quantiative vars, Euclidian distance) • Similarity measure srs (=1 if same object, <1 otherwise) – Ex: correlation • Dissimilarity measure δrs (=0 if same object, >0 otherwise) – Ex: euclidian distance 732 A 44 Programming in R

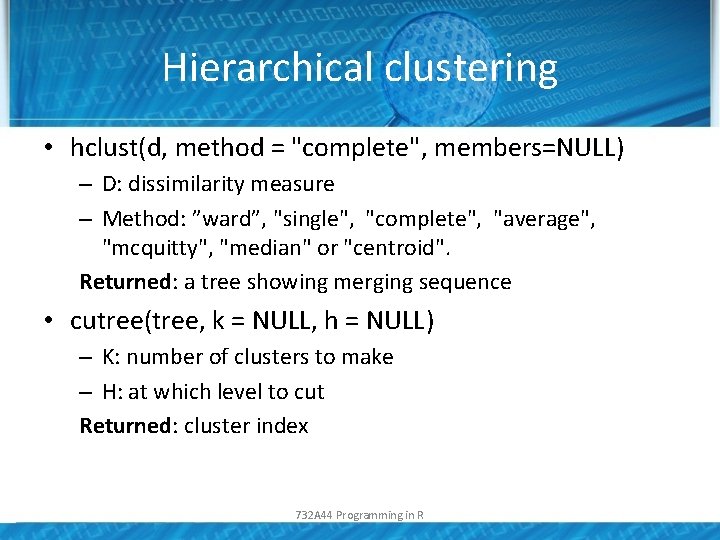
Hierarchical clustering • hclust(d, method = "complete", members=NULL) – D: dissimilarity measure – Method: ”ward”, "single", "complete", "average", "mcquitty", "median" or "centroid". Returned: a tree showing merging sequence • cutree(tree, k = NULL, h = NULL) – K: number of clusters to make – H: at which level to cut Returned: cluster index 732 A 44 Programming in R

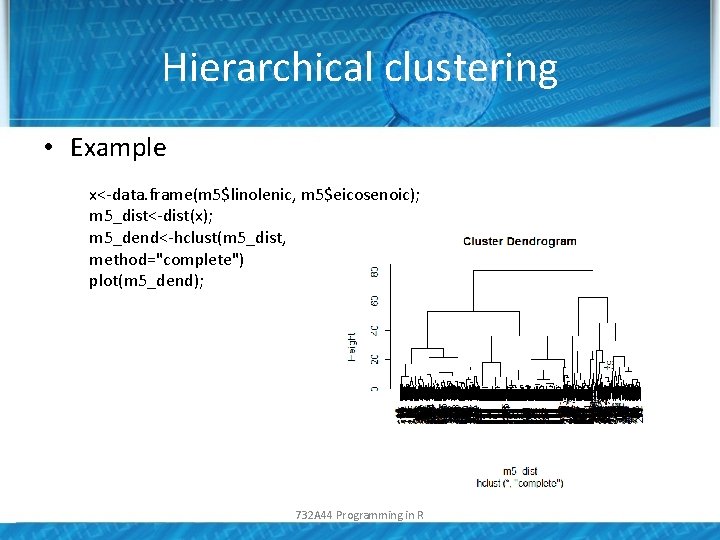
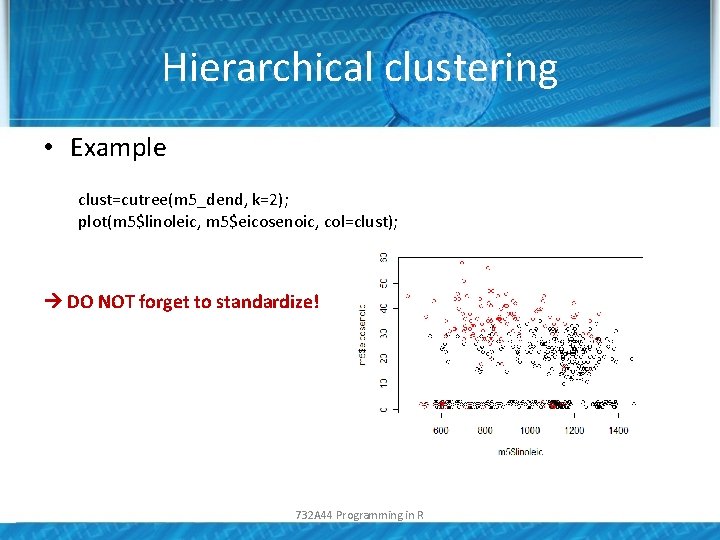
Hierarchical clustering • Example x<-data. frame(m 5$linolenic, m 5$eicosenoic); m 5_dist<-dist(x); m 5_dend<-hclust(m 5_dist, method="complete") plot(m 5_dend); 732 A 44 Programming in R

Hierarchical clustering • Example clust=cutree(m 5_dend, k=2); plot(m 5$linoleic, m 5$eicosenoic, col=clust); DO NOT forget to standardize! 732 A 44 Programming in R

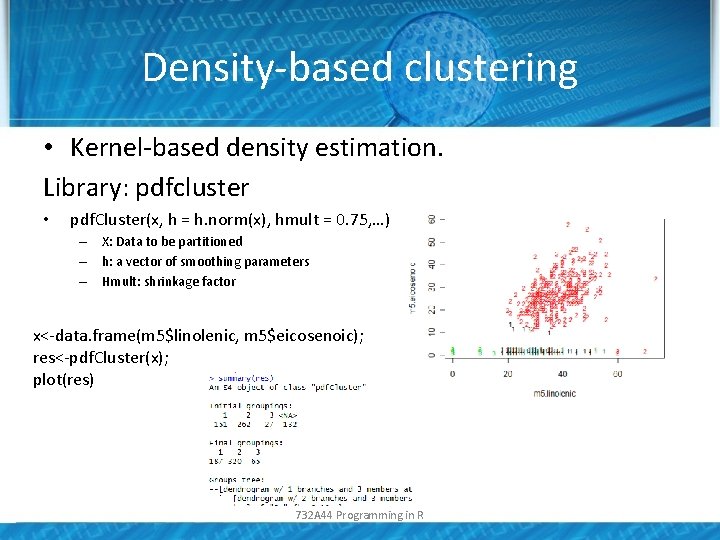
Density-based clustering • Kernel-based density estimation. Library: pdfcluster • pdf. Cluster(x, h = h. norm(x), hmult = 0. 75, …) – X: Data to be partitioned – h: a vector of smoothing parameters – Hmult: shrinkage factor x<-data. frame(m 5$linolenic, m 5$eicosenoic); res<-pdf. Cluster(x); plot(res) 732 A 44 Programming in R

Reference http: //cran. r-project. org/doc/contrib/Yanchang. Zhaorefcard-data-mining. pdf 732 A 44 Programming in R