Lecture 4 Sequence alignment and searching Sequence alignment









- Slides: 37

Lecture 4 Sequence alignment and searching

Sequence alignment

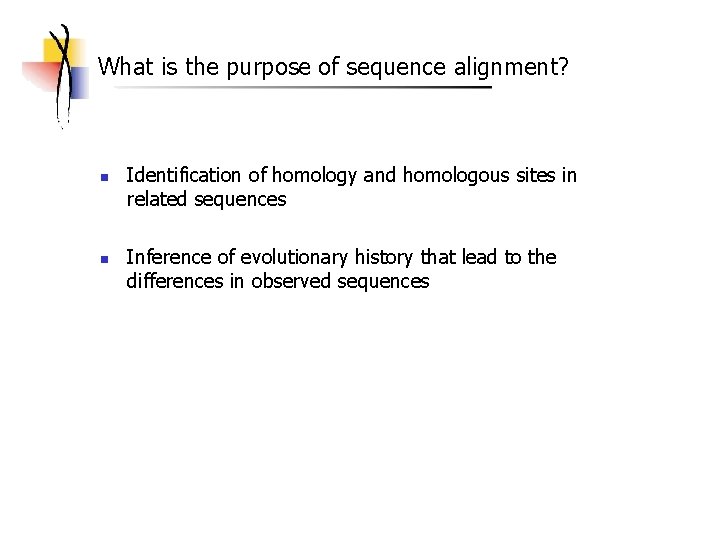
What is the purpose of sequence alignment? n n Identification of homology and homologous sites in related sequences Inference of evolutionary history that lead to the differences in observed sequences

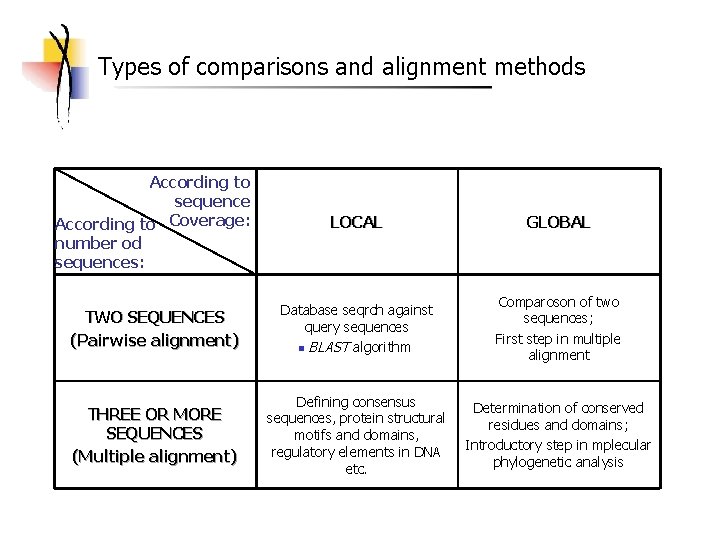
Types of comparisons and alignment methods According to sequence According to Coverage: number od sequences: LOCAL GLOBAL TWO SEQUENCES (Pairwise alignment) Database seqrch against query sequences n BLAST algorithm Comparoson of two sequences; First step in multiple alignment THREE OR MORE SEQUENCES (Multiple alignment) Defining consensus sequences, protein structural motifs and domains, regulatory elements in DNA etc. Determination of conserved residues and domains; Introductory step in mplecular phylogenetic analysis

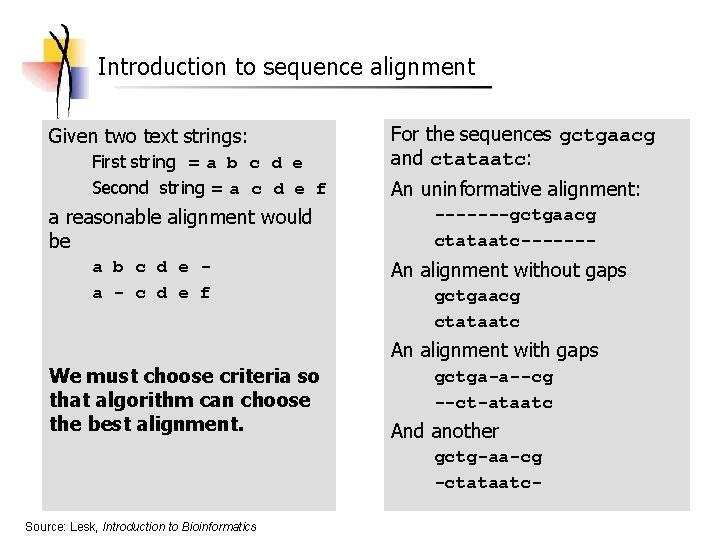
Introduction to sequence alignment Given two text strings: First string = a b c d e Second string = a c d e f a reasonable alignment would be a b c d e a - c d e f For the sequences gctgaacg and ctataatc: An uninformative alignment: -------gctgaacg ctataatc------- An alignment without gaps gctgaacg ctataatc An alignment with gaps We must choose criteria so that algorithm can choose the best alignment. gctga-a--cg --ct-ataatc And another gctg-aa-cg -ctataatc- Source: Lesk, Introduction to Bioinformatics

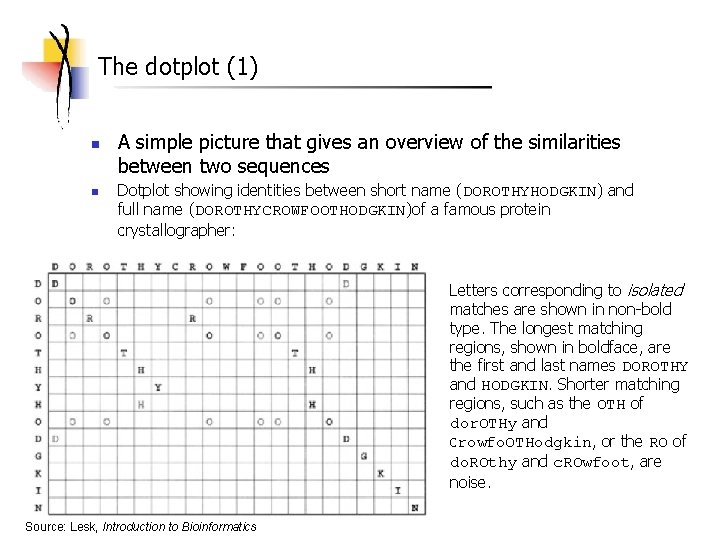
The dotplot (1) n n A simple picture that gives an overview of the similarities between two sequences Dotplot showing identities between short name (DOROTHYHODGKIN) and full name (DOROTHYCROWFOOTHODGKIN)of a famous protein crystallographer: Letters corresponding to isolated matches are shown in non-bold type. The longest matching regions, shown in boldface, are the first and last names DOROTHY and HODGKIN. Shorter matching regions, such as the OTH of dor. OTHy and crowfo. OTHodgkin, or the RO of do. ROthy and c. ROwfoot, are noise. Source: Lesk, Introduction to Bioinformatics

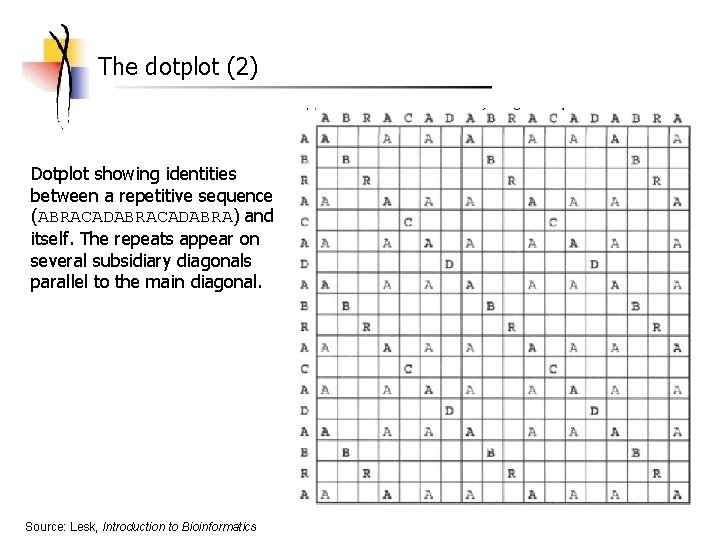
The dotplot (2) Dotplot showing identities between a repetitive sequence (ABRACADABRA) and itself. The repeats appear on several subsidiary diagonals parallel to the main diagonal. Source: Lesk, Introduction to Bioinformatics

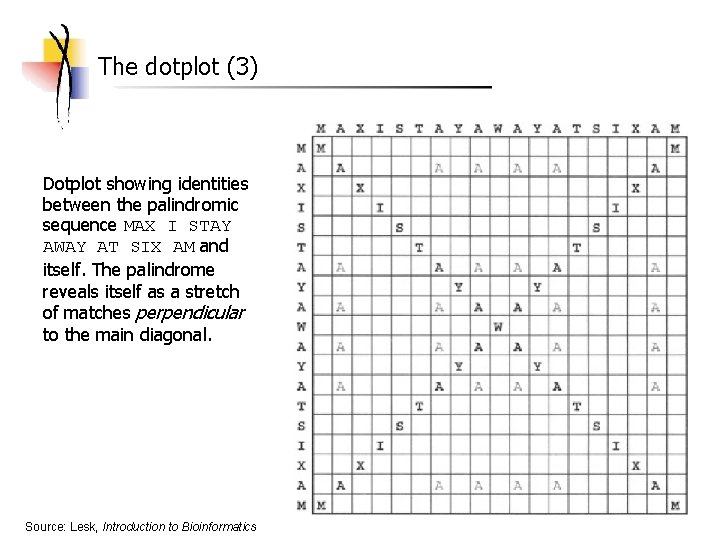
The dotplot (3) Dotplot showing identities between the palindromic sequence MAX I STAY AWAY AT SIX AM and itself. The palindrome reveals itself as a stretch of matches perpendicular to the main diagonal. Source: Lesk, Introduction to Bioinformatics

The dotplot (4) Source: Lesk, Introduction to Bioinformatics

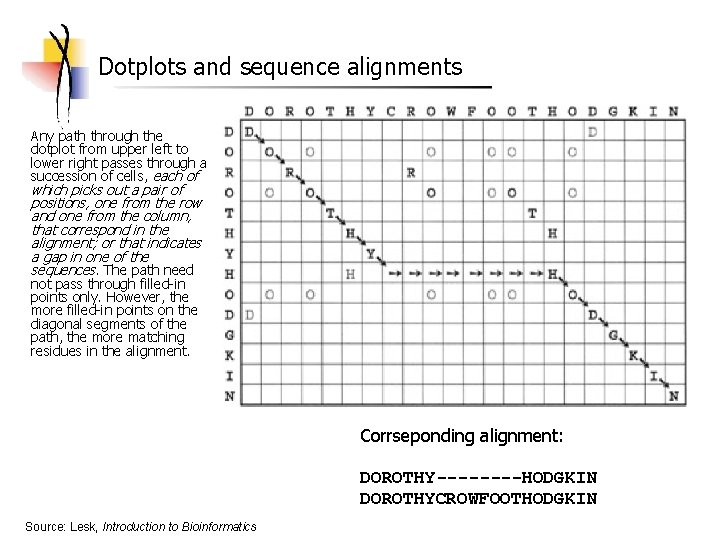
Dotplots and sequence alignments Any path through the dotplot from upper left to lower right passes through a succession of cells, each of which picks out a pair of positions, one from the row and one from the column, that correspond in the alignment; or that indicates a gap in one of the sequences. The path need not pass through filled-in points only. However, the more filled-in points on the diagonal segments of the path, the more matching residues in the alignment. Corrseponding alignment: DOROTHY----HODGKIN DOROTHYCROWFOOTHODGKIN Source: Lesk, Introduction to Bioinformatics

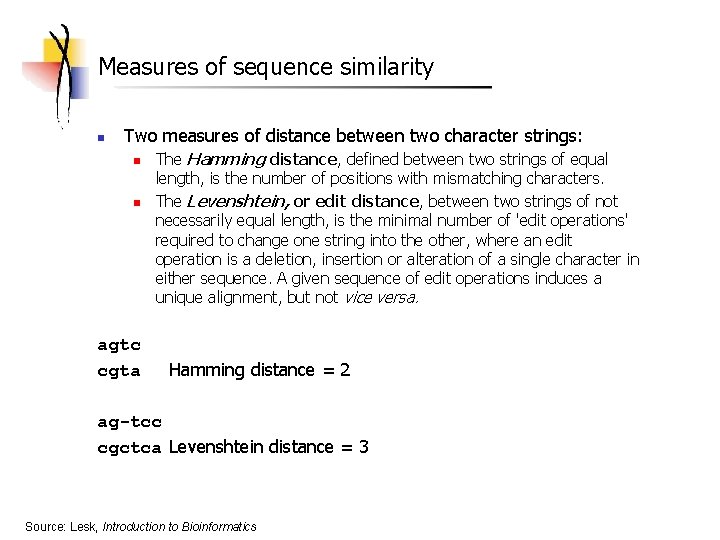
Measures of sequence similarity n Two measures of distance between two character strings: n n agtc cgta The Hamming distance, defined between two strings of equal length, is the number of positions with mismatching characters. The Levenshtein, or edit distance, between two strings of not necessarily equal length, is the minimal number of 'edit operations' required to change one string into the other, where an edit operation is a deletion, insertion or alteration of a single character in either sequence. A given sequence of edit operations induces a unique alignment, but not vice versa. Hamming distance = 2 ag-tcc cgctca Levenshtein distance = 3 Source: Lesk, Introduction to Bioinformatics

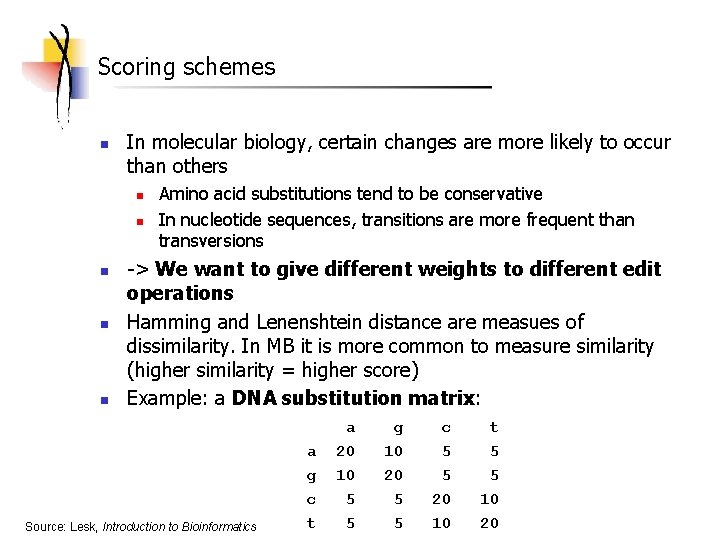
Scoring schemes n In molecular biology, certain changes are more likely to occur than others n n n Amino acid substitutions tend to be conservative In nucleotide sequences, transitions are more frequent than transversions -> We want to give different weights to different edit operations Hamming and Lenenshtein distance are measues of dissimilarity. In MB it is more common to measure similarity (higher similarity = higher score) Example: a DNA substitution matrix: Source: Lesk, Introduction to Bioinformatics a g c t a 20 10 5 5 g 10 20 5 5 c 5 5 20 10 t 5 5 10 20

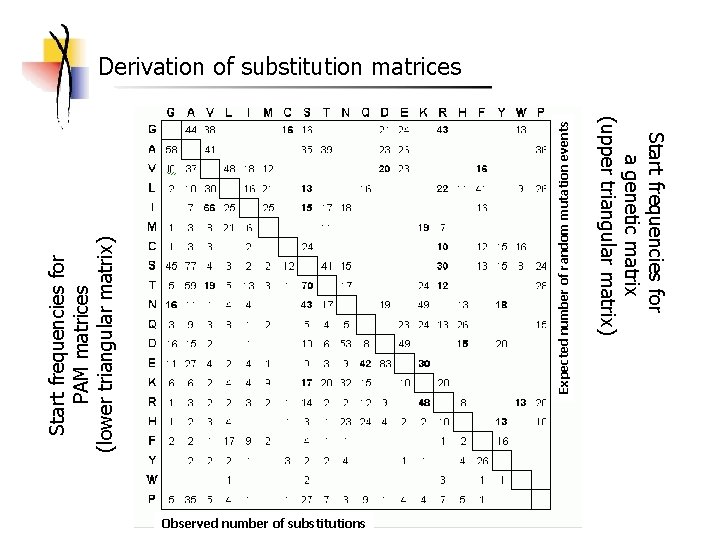
Observed number of substitutions Start frequencies for a genetic matrix (upper triangular matrix) Expected number of random mutation events Start frequencies for PAM matrices (lower triangular matrix) Derivation of substitution matrices

PAM Matrices n n n First substitution matrices widely used Based on the point-accepted-mutation (PAM) model of evolution (Dayhoff, 1978) PAMs are relative measures of evolutionary distance n n 1 PAM = 1 accepted mutation per 100 AAs Does not mean that after 100 PAMs every AA will be different? Why or why not? To view and compare different substitution matrices, visit http: //eta. embl-heidelberg. de: 8000/misc/mat/

PAM Matrices n n Derived from global alignments of closely related sequences. Matrices for greater evolutionary distances are extrapolated from those for lesser ones. The number with the matrix (PAM 40, PAM 100) refers to the evolutionary distance; greater numbers are greater distances. Does not take into account different evolutionary rates between conserved and non-conserved regions.

BLOSUM Matrices n n n n Henikoff, S. & Henikoff J. G. (1992) Use blocks of protein sequence fragments from different families (the BLOCKS database) Amino acid pair frequencies calculated by summing over all possible pairs in block Different evolutionary distances are incorporated into this scheme with a clustering procedure (identity over particular threshold = same cluster) Similar idea to PAM matrices Probabilities estimated from blocks of sequence fragments Blocks represent structurally conserved regions

BLOSUM Matrices n n Target frequencies are identified directly instead of extrapolation. Sequences more than x% identitical within the block where substitutions are being counted, are grouped together and treated as a single sequence n n BLOSUM 50 : >= 50% identity BLOSUM 62 : >= 62 % identity To view and compare different substitution matrices, visit http: //eta. embl-heidelberg. de: 8000/misc/mat/

BLOSUM Matrices n n Derived from local, ungapped alignments of distantly related sequences All matrices are directly calculated; no extrapolations are used – no explicit model The number after the matrix (BLOSUM 62) refers to the minimum percent identity of the blocks used to construct the matrix; greater numbers are lesser distances. The BLOSUM series of matrices generally perform better than PAM matrices for local similarity searches (Proteins 17: 49).

How do you prefer it: done right or done fast? n Exact methods - the result is guaranteed to be (mathematically) optimal n Pairwise alignment n n n Multiple alignment n n Needleman-Wunsch (global) Smith-Waterman (local) Rarely used; quickly becomes computationally intractable with increasing number of sequences and/or their length Heuristic methods: make some assumptions that hold most, but not all of the time n n Pairwise alignment – BLAST Multiple alignment – all widely used multiple alignment programs (Clustal. W, TCofee…)

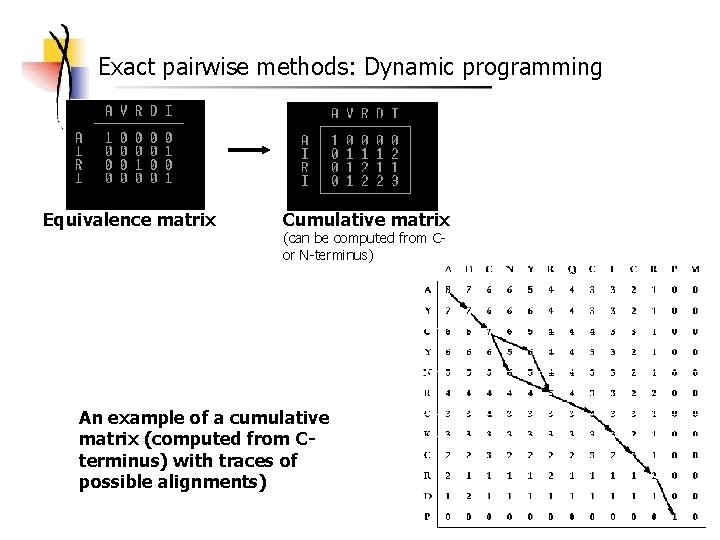
Exact pairwise methods: Dynamic programming Equivalence matrix Cumulative matrix (can be computed from Cor N-terminus) An example of a cumulative matrix (computed from Cterminus) with traces of possible alignments)

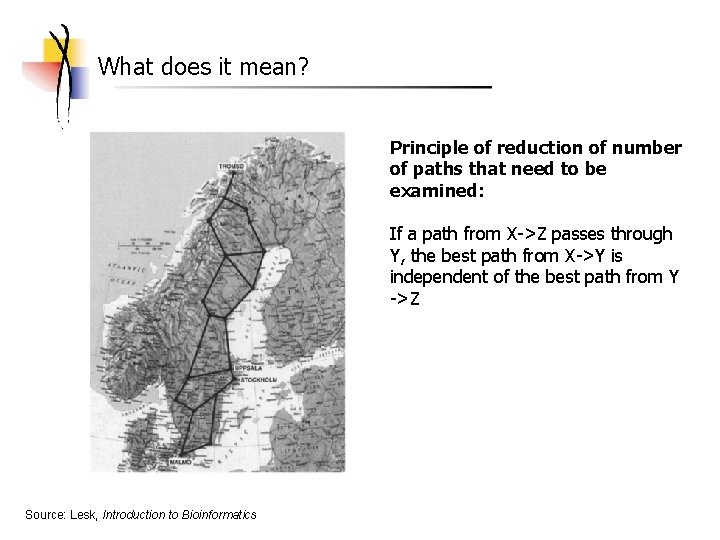
What does it mean? Principle of reduction of number of paths that need to be examined: If a path from X->Z passes through Y, the best path from X->Y is independent of the best path from Y ->Z Source: Lesk, Introduction to Bioinformatics

BLAST – the workhorse of bioinformatcs http: //www. ncbi. nlm. nih. gov/BLAST n n BLAST = Basic local alignment search tool When you have a nucleotide or protein sequence that you want to search against sequence databases n n n to determine what the sequence is to find related sequences (homologs) Bioinformatics researchers use standalone version of BLAST, run from the command line or other programs

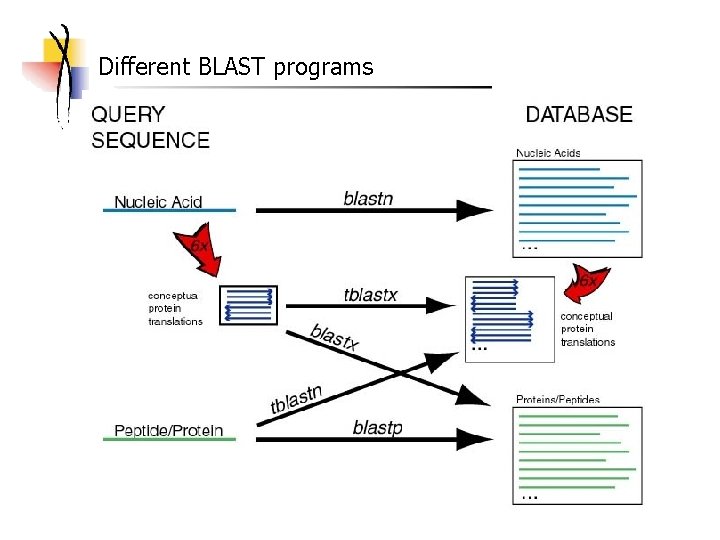
Different BLAST programs

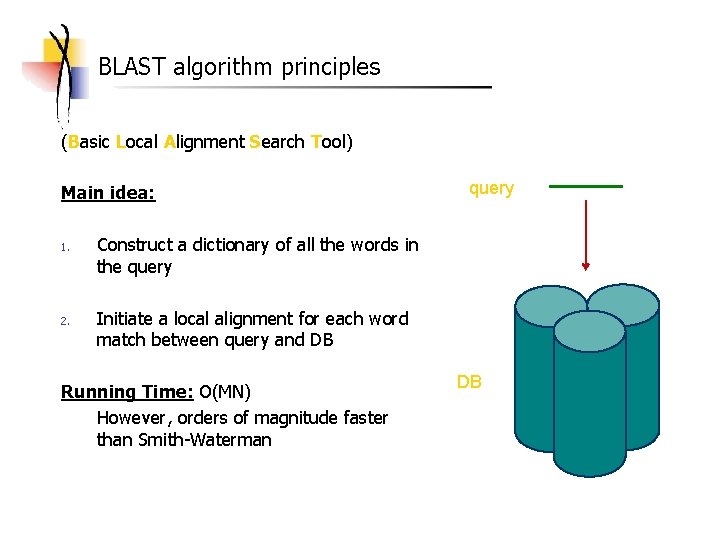
BLAST algorithm principles (Basic Local Alignment Search Tool) Main idea: 1. 2. query Construct a dictionary of all the words in the query Initiate a local alignment for each word match between query and DB Running Time: O(MN) However, orders of magnitude faster than Smith-Waterman DB

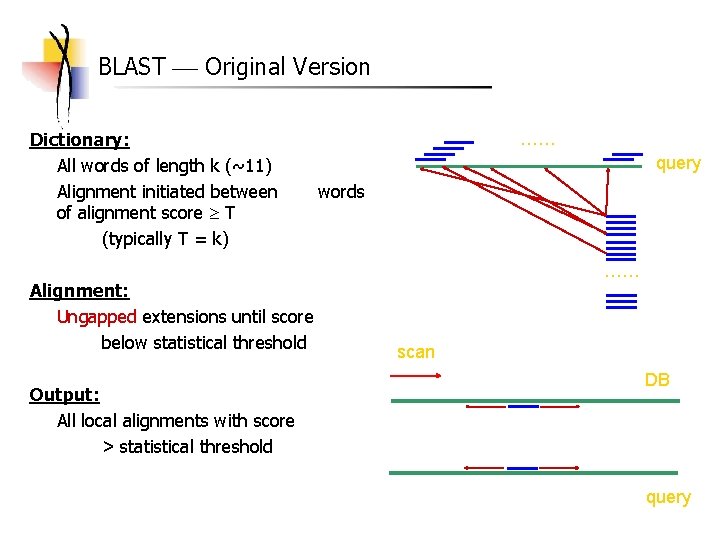
BLAST Original Version Dictionary: All words of length k (~11) Alignment initiated between of alignment score T (typically T = k) Alignment: Ungapped extensions until score below statistical threshold Output: All local alignments with score > statistical threshold …… query words …… scan DB query

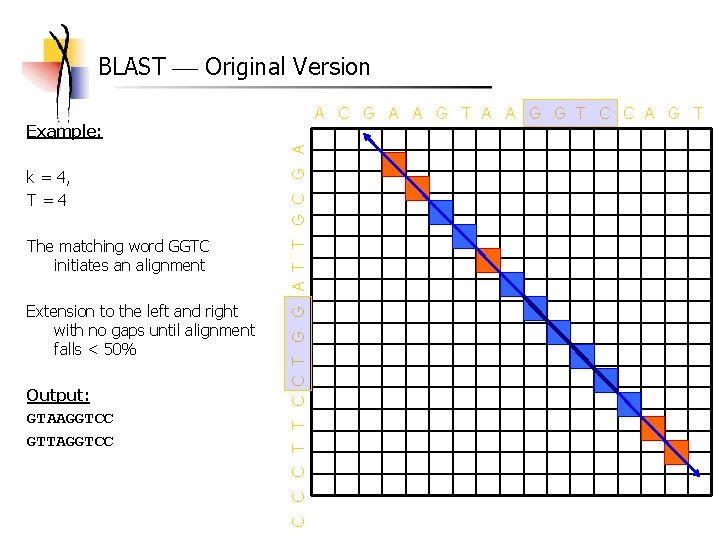
BLAST Original Version A C G A A G T A A G G T C C A G T k = 4, T=4 The matching word GGTC initiates an alignment Extension to the left and right with no gaps until alignment falls < 50% Output: GTAAGGTCC GTTAGGTCC C T T C C T G G A T T G C G A Example:

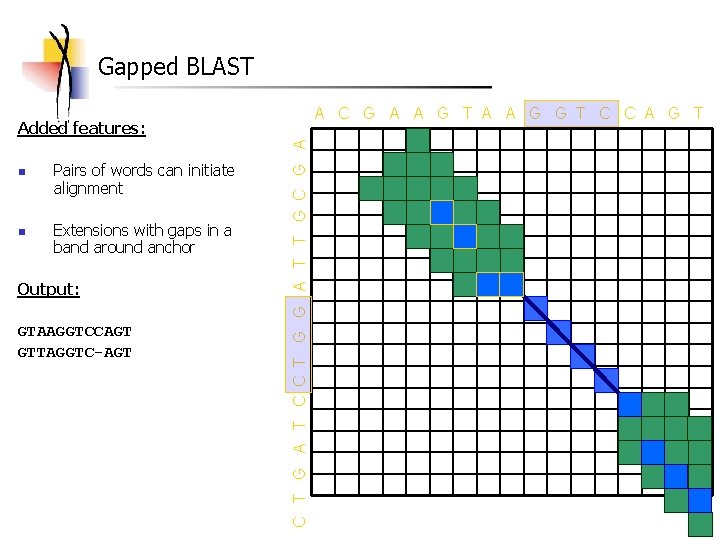
Gapped BLAST A C G A A G T A A G G T C C A G T n n Pairs of words can initiate alignment Extensions with gaps in a band around anchor Output: GTAAGGTCCAGT GTTAGGTC-AGT C T G A T C C T G G A T T G C G A Added features:

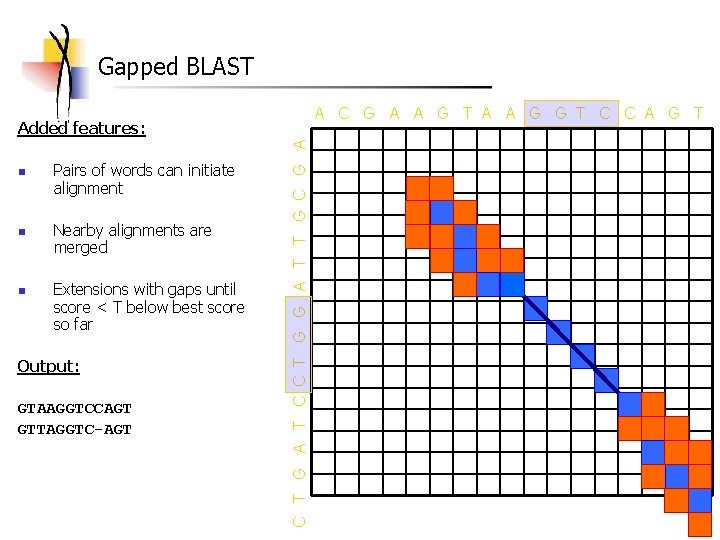
Gapped BLAST A C G A A G T A A G G T C C A G T n n n Pairs of words can initiate alignment Nearby alignments are merged Extensions with gaps until score < T below best score so far Output: GTAAGGTCCAGT GTTAGGTC-AGT C T G A T C C T G G A T T G C G A Added features:

BLAST: Example of sequence search Query sequence or accession number Sequence database to search

BLAST: Example of sequence search Please come back later

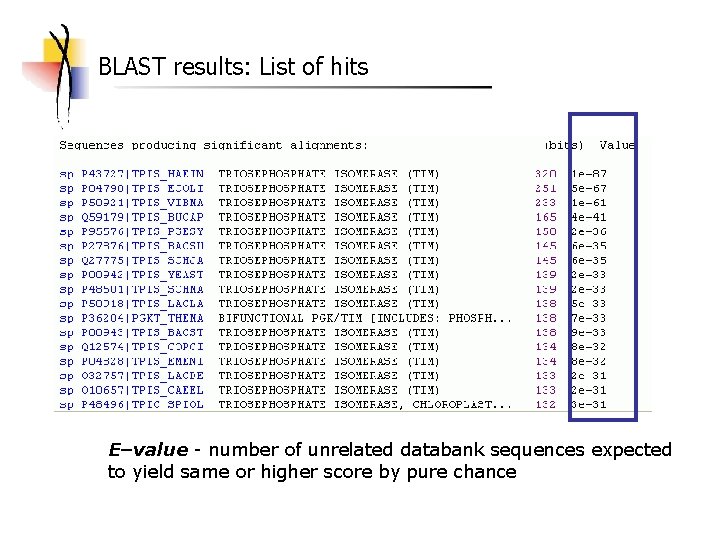
BLAST results: List of hits E–value - number of unrelated databank sequences expected to yield same or higher score by pure chance

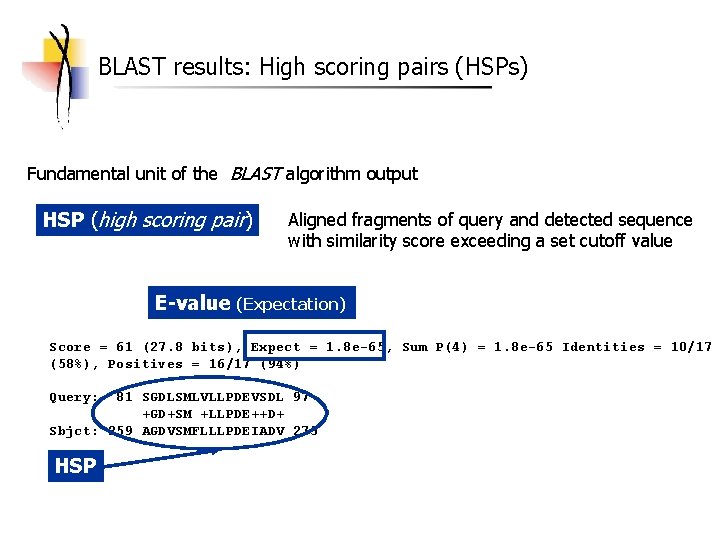
BLAST results: High scoring pairs (HSPs) Fundamental unit of the BLAST algorithm output HSP (high scoring pair) Aligned fragments of query and detected sequence with similarity score exceeding a set cutoff value E-value (Expectation) Score = 61 (27. 8 bits), Expect = 1. 8 e-65, Sum P(4) = 1. 8 e-65 Identities = 10/17 (58%), Positives = 16/17 (94%) Query: 81 SGDLSMLVLLPDEVSDL 97 +GD+SM +LLPDE++D+ Sbjct: 259 AGDVSMFLLLPDEIADV 275 HSP

BLAST Confidence measures Score and bit-score : depend on scoring method E-value (Expect value) : number of unrelated database sequences expected to yield same or higher score by pure chance P-value (Probability) : probability that database yields by pure chance at least one alignment with same or higer score The Expect value (E) is a parameter that describes the number of hits one can "expect" to see just by chance when searching a database of a particular size. It decreases exponentially with the Score (S) that is assigned to a match between two sequences. Essentially, the E value describes the random background noise that exists for matches between sequences. The Expect value is used as a convenient way to create a significance threshold for reporting results. When the Expect value is increased from the default value of 10, a larger list with more low-scoring hits can be reported. (E-value approching zero => significant alignment. Less than 0, 01 = almost always found to be homologous; 1 e-10 for nucleotide searches of 1 e 4 for protein searches= frequently related) In BLAST 2. 0, the Expect value is also used instead of the P value (probability) to report the significance of matches. For example, an E value of 1 assigned to a hit can be interpreted as meaning that in a database of the current size one might expect to see 1 match with a similar score simply by chance.

Be careful when comparing E- or P-values from different searches Comparison is only meaningful for different query sequences searched agains the same database with the same BLAST parameters

Variants of BLAST n MEGABLAST: n Optimized to align very similar sequences n n n PSI-BLAST: n n BLAST produces many hits Those are aligned, and a pattern is extracted Pattern is used for next search; above steps iterated WU-BLAST: (Washington University BLAST) n n Works best when k = 4 i 16 Linear gap penalty Optimized, added features Blast. Z n n Combines BLAST/Pattern. Hunter methodology Good for local alignments of non-coding (regulatory) regions

IMPORTANT: When NOT to use BLAST n n n A typical situation: you have a piece of human DNA sequence and want to extend it or find what gene it belongs to. In other words, you want an exact or near-exact match to a sequence that is part of an assembled genome. Genome browsers are equipped with very fast algorithms for finding near-exact matches in genomic sequences: n BLAT (with UCSC Genome Browser) n n Highly recommended: the BLAT paper (Kent WJ (2003) Genome Res 12: 656 -64) - it is famous for its unorthodox writing style SSAHA (with Ensembl)

BLAT: BLAST-like alignment tool n Differences with respect to BLAST: n Dictionary of the DB (BLAST: query) n Initiate alignment on any # of matching hits (BLAST: 1 or 2 hits) n More aggressive stitching-together of alignments n Special code to align mature m. RNA to DNA n Result: very efficient for: n n Aligning many sequences to a DB Aligning two genomes