Lecture 4 Polymerase Chain Reaction PCR Dr Shah



- Slides: 19

Lecture 4: Polymerase Chain Reaction (PCR) Dr. Shah Rukh Abbas ASAB, NUST

Polymerase Chain Reaction • Polymerase: DNA polymerase – DNA polymerase duplicates DNA – Before a cell divides, its DNA must be duplicated • Chain Reaction: The product of a reaction is used to amplify the same reaction – Results in rapid increase in the product

Polymerase Chain Reaction (PCR) • • • PCR performs the chemistry of DNA duplication in vitro Numerous PCR applications make this process a staple in most biology laboratories Understanding properties of DNA polymerases helps understanding PCR

Discovery • PCR was discovered by Kary Mullis – On a long motorcycle drive – According to Mullis, he was driving his vehicle late night with his girlfriend – Mentally visualized the process • Nobel Prize in Chemistry – 1993

DNA polymerase • Duplicates DNA • Necessary for reproduction of new cells • More than one DNA polymerases exist in different organisms

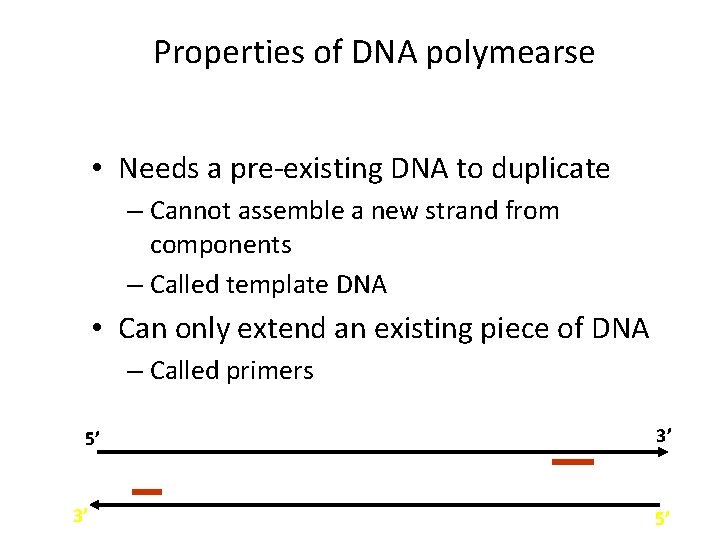
Properties of DNA polymearse • Needs a pre-existing DNA to duplicate – Cannot assemble a new strand from components – Called template DNA • Can only extend an existing piece of DNA – Called primers 5’ 3’ 3’ 5’

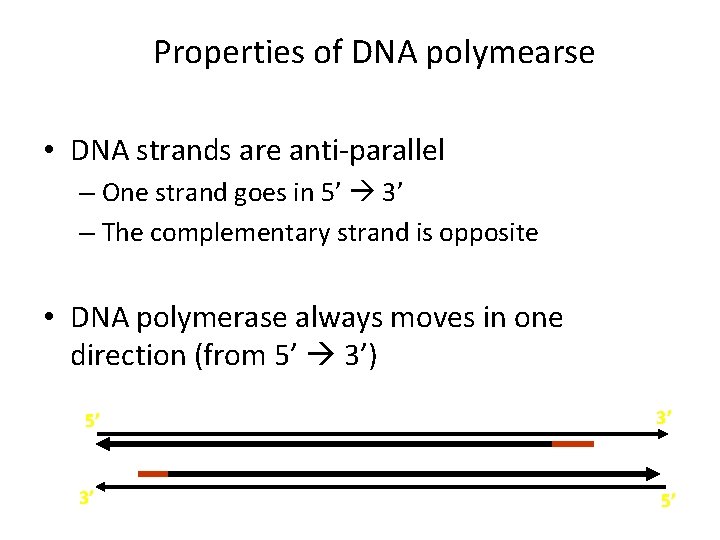
Properties of DNA polymearse • DNA strands are anti-parallel – One strand goes in 5’ 3’ – The complementary strand is opposite • DNA polymerase always moves in one direction (from 5’ 3’) 5’ 3’ 3’ 5’

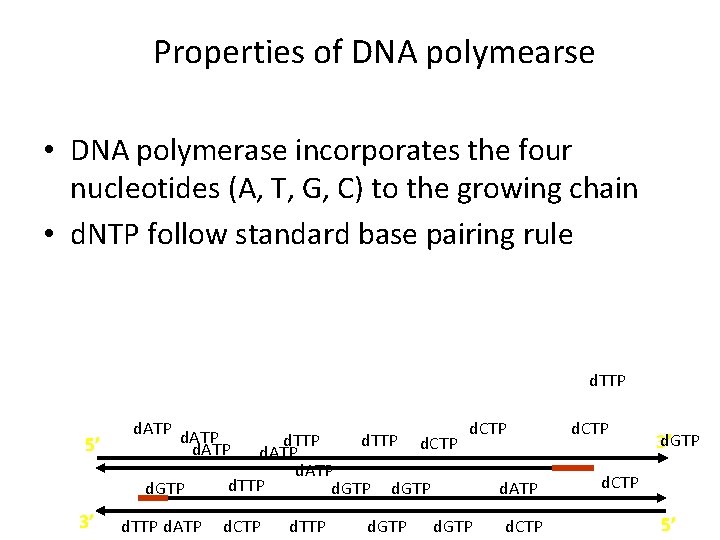
Properties of DNA polymearse • DNA polymerase incorporates the four nucleotides (A, T, G, C) to the growing chain • d. NTP follow standard base pairing rule d. TTP 5’ d. ATP d. GTP 3’ d. TTP d. ATP d. TTP d. CTP d. ATP d. TTP d. GTP d. CTP d. GTP d. ATP d. CTP d. GTP 3’ d. CTP 5’

Properties of DNA polymearse • The newly generated DNA strands serve as template DNA for the next cycle • PCR is very sensitive • Widely used

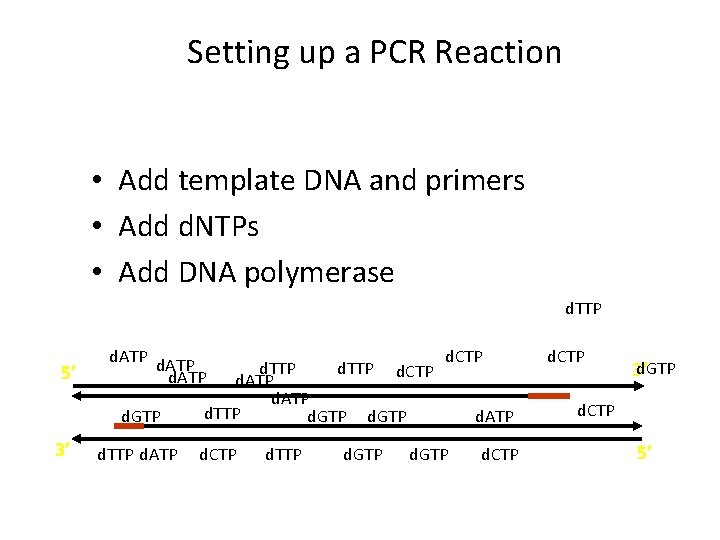
Setting up a PCR Reaction • Add template DNA and primers • Add d. NTPs • Add DNA polymerase d. TTP 5’ d. ATP d. GTP 3’ d. TTP d. ATP d. TTP d. CTP d. ATP d. TTP d. GTP d. CTP d. GTP d. ATP d. CTP d. GTP 3’ d. CTP 5’

Properties of DNA polymearse • DNA polymerase needs Mg++ as cofactor • Each DNA polymerase works best under optimal temperature, p. H and salt concentration • PCR buffer provides optimal p. H and salt condition

Taq DNA polymerase • Derived from Thermus aquaticus • Heat stable DNA polymerase • Ideal temperature 72 C

Thermal Cycling • A PCR machine controls temperature • Typical PCR go through three steps – Denaturation – Annealing – Extension

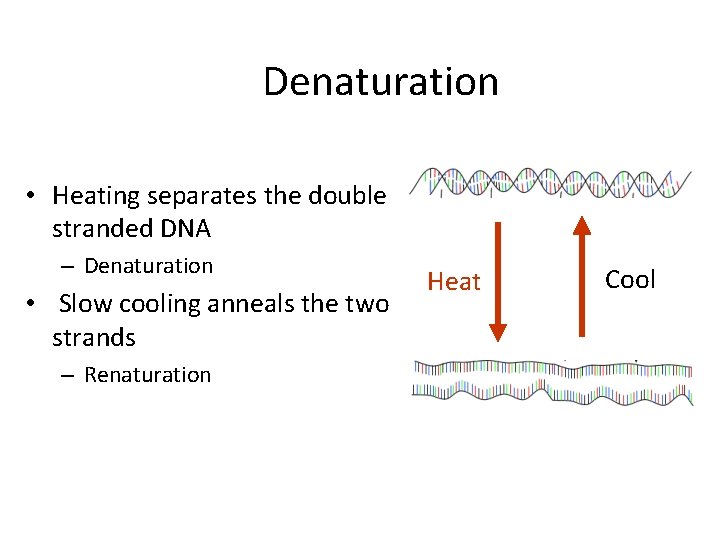
Denaturation • Heating separates the double stranded DNA – Denaturation • Slow cooling anneals the two strands – Renaturation Heat Cool

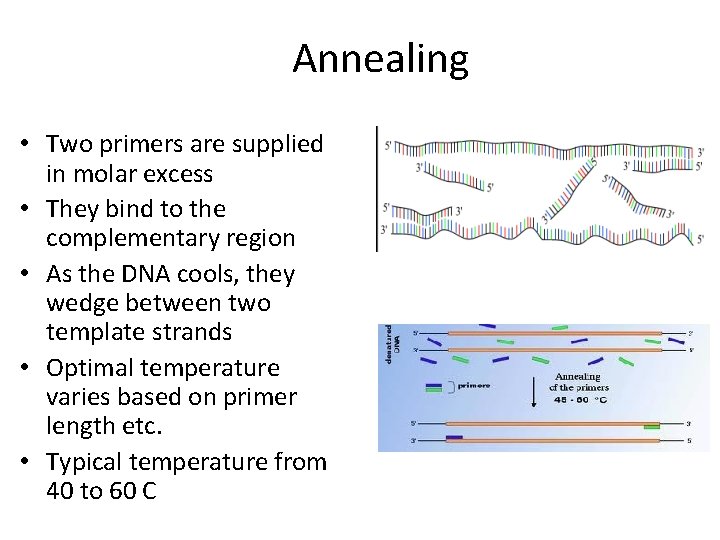
Annealing • Two primers are supplied in molar excess • They bind to the complementary region • As the DNA cools, they wedge between two template strands • Optimal temperature varies based on primer length etc. • Typical temperature from 40 to 60 C

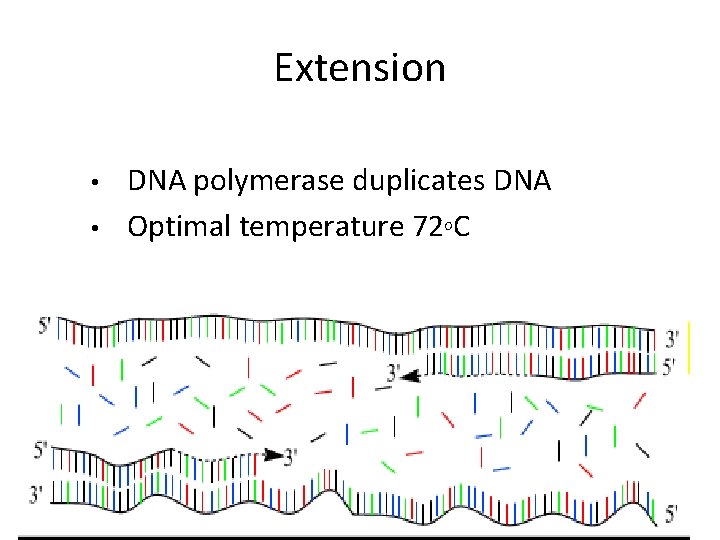
Extension • • DNA polymerase duplicates DNA Optimal temperature 72 OC

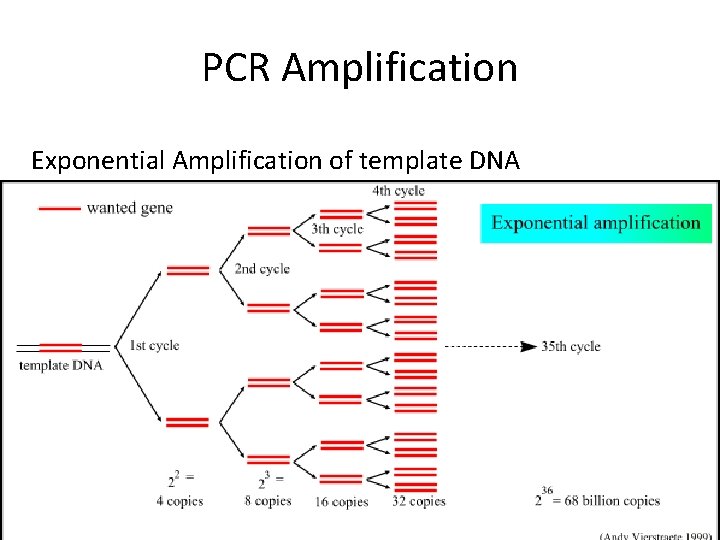
PCR Amplification Exponential Amplification of template DNA

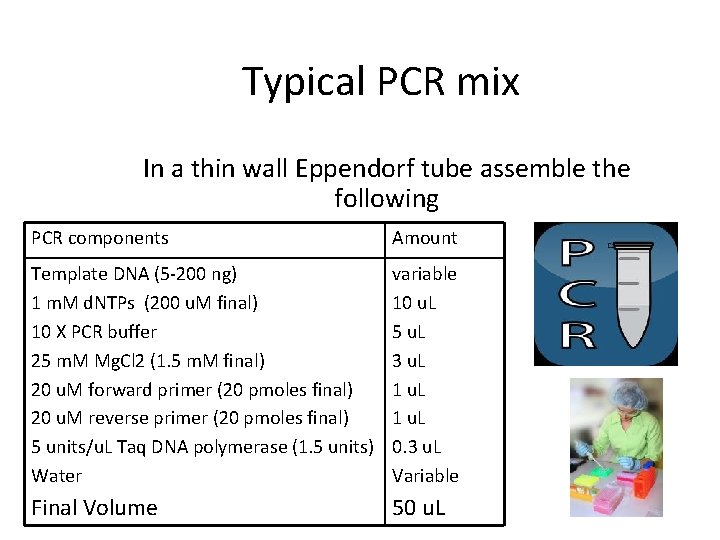
Typical PCR mix In a thin wall Eppendorf tube assemble the following PCR components Amount Template DNA (5 -200 ng) 1 m. M d. NTPs (200 u. M final) 10 X PCR buffer 25 m. M Mg. Cl 2 (1. 5 m. M final) 20 u. M forward primer (20 pmoles final) 20 u. M reverse primer (20 pmoles final) 5 units/u. L Taq DNA polymerase (1. 5 units) Water variable 10 u. L 5 u. L 3 u. L 1 u. L 0. 3 u. L Variable Final Volume 50 u. L

Applications • Revolutionized how we study biology – Recombinant DNA Technology – Research – Diagnostics – Forensics