Lecture 24 DNA replication G C A T


- Slides: 22

Lecture 24: DNA replication G C A T T A 1 nm C G C A T G C T A A T T A G A Figure 16. 7 a, c 3. 4 nm G C 0. 34 nm T (c) Space-filling model

Lecture Outline • • Overview of DNA replication DNA has polarity (5’P, 3’OH) Rules of Replication Details of the replication machine – Proofreading – Mutation – Special problems at the ends of chromosomes G C A T T A 1 nm C G C 3. 4 nm G A T G C T A A T T A G A C T 0. 34 nm

Review • Watson-Crick model of DNA structure • Base composition ratios – A=T; G=C • Radioactive labeling experiments show that – DNA is the genetic material – Replication is semi-conservative G C A T T A 1 nm C G C 3. 4 nm G A T G C T A A T T A G A C T 0. 34 nm

Chromosomes • Bacterial chromosomes tend to be circular, eukaryotic chromosomes linear • DNA plus associated proteins = chromatin – Euchromatin vs heterochromatin


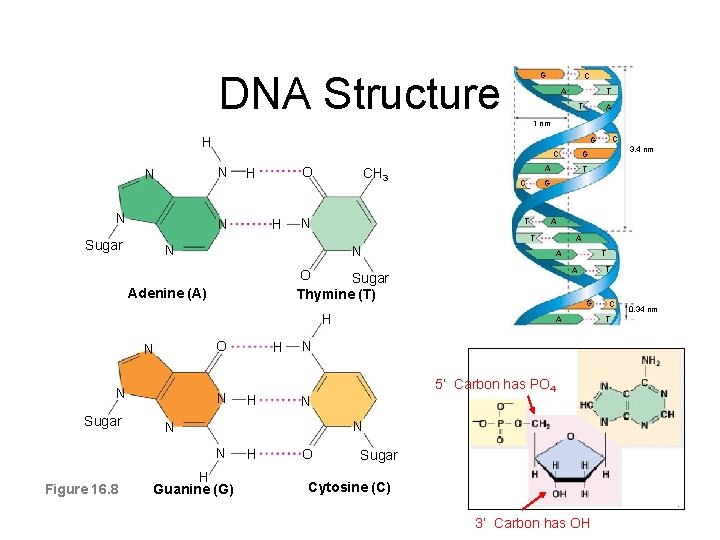
DNA Structure G C A T T A 1 nm H C N N N O H N H CH 3 N T G C T N A N O N Sugar N H T G A N 5’ Carbon has PO 4 N N Figure 16. 8 H T A H N A A O Sugar Thymine (T) Adenine (A) H Guanine (G) H O 3. 4 nm G A T Sugar C G Sugar Cytosine (C) 3’ Carbon has OH C T 0. 34 nm


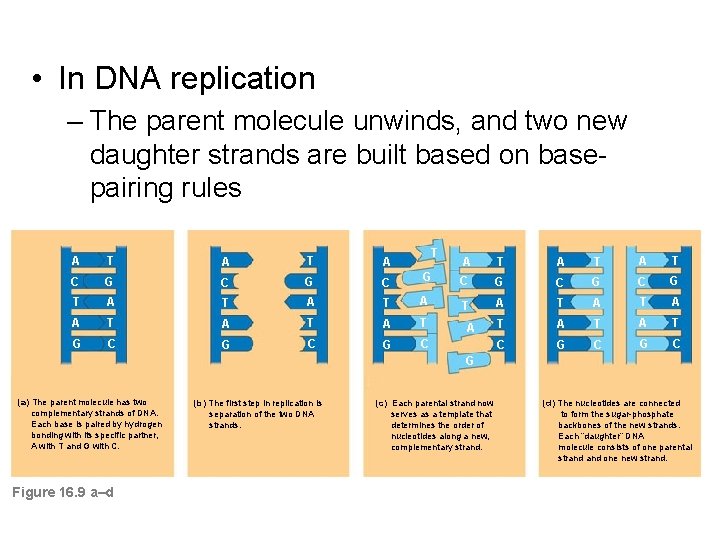
• In DNA replication – The parent molecule unwinds, and two new daughter strands are built based on basepairing rules T A T A C G C T A T A T G C G C G A T A T C G C G T A T A T A T C G C A G (a) The parent molecule has two complementary strands of DNA. Each base is paired by hydrogen bonding with its specific partner, A with T and G with C. Figure 16. 9 a–d (b) The first step in replication is separation of the two DNA strands. (c) Each parental strand now serves as a template that determines the order of nucleotides along a new, complementary strand. (d) The nucleotides are connected to form the sugar-phosphate backbones of the new strands. Each “daughter” DNA molecule consists of one parental strand one new strand.

Replication overview • Must maintain integrity of the DNA sequence through successive rounds of replication – (think of the game “telephone”) • Need to: – unwind DNA, add an RNA primer, find an appropriate base, add it to the growing DNA fragment, proofread, remove the initial primer, fill in the gap with DNA, ligate fragments together • All of this is fast, about 100 bp/second animation from DNAi

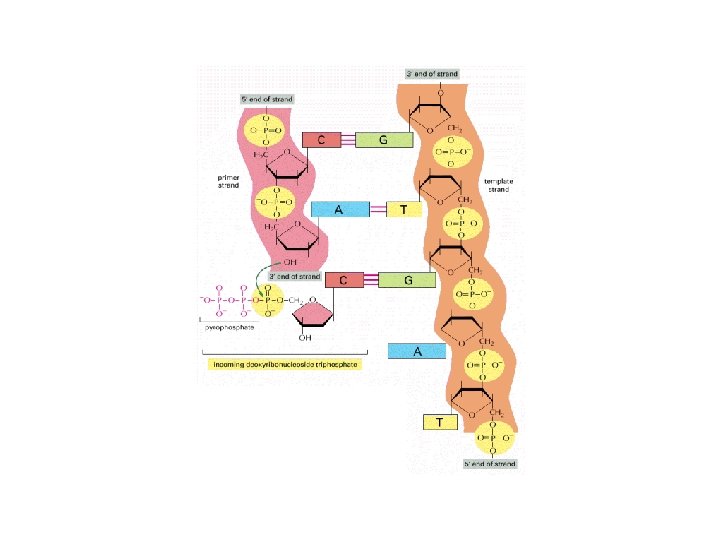
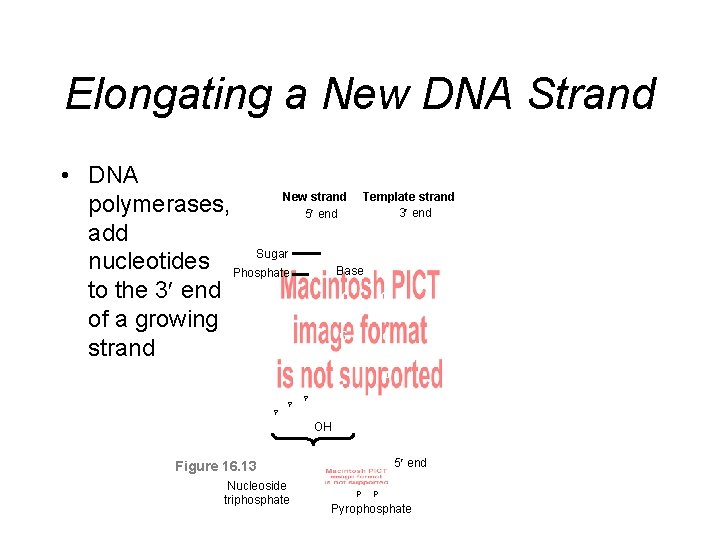
Elongating a New DNA Strand • DNA New strand Template strand polymerases, 3 end 5 end add Sugar A T nucleotides Phosphate Base to the 3 end C G of a growing G C strand A P P T P C OH 5 end Figure 16. 13 Nucleoside triphosphate P P Pyrophosphate

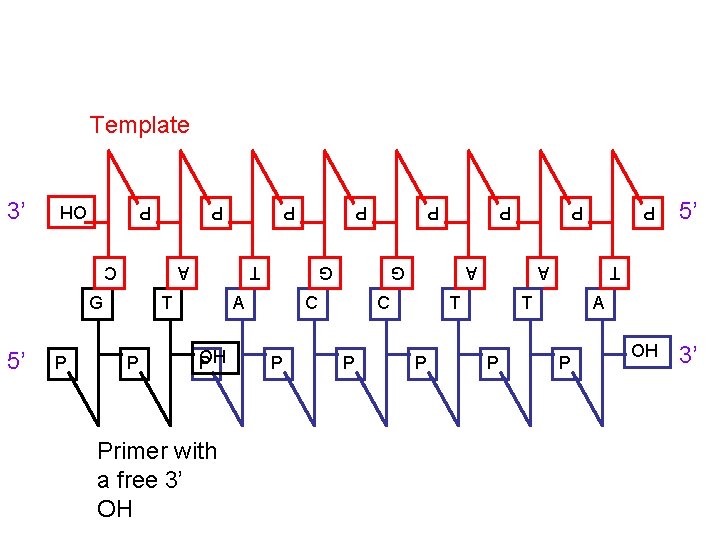
Template G A P G T OH P A C Primer with a free 3’ OH P C P A P OH P C T P 5’ OH 3’ T P P A P 5’ T P G P 3’ A P

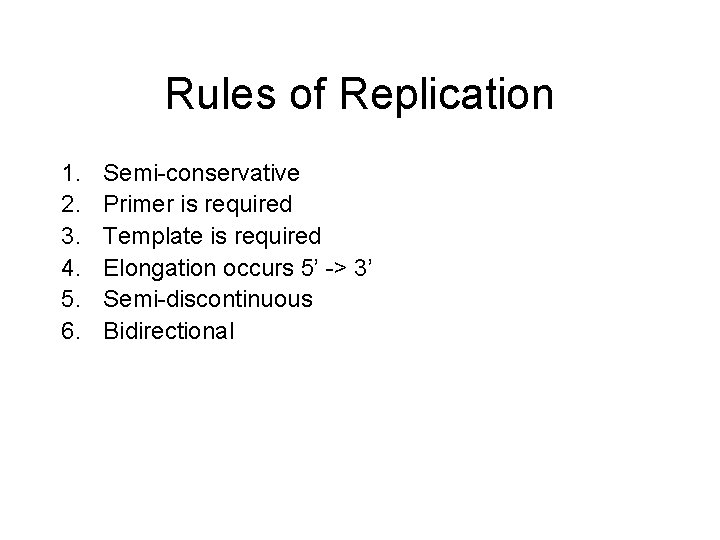
Rules of Replication 1. 2. 3. 4. 5. 6. Semi-conservative Primer is required Template is required Elongation occurs 5’ -> 3’ Semi-discontinuous Bidirectional

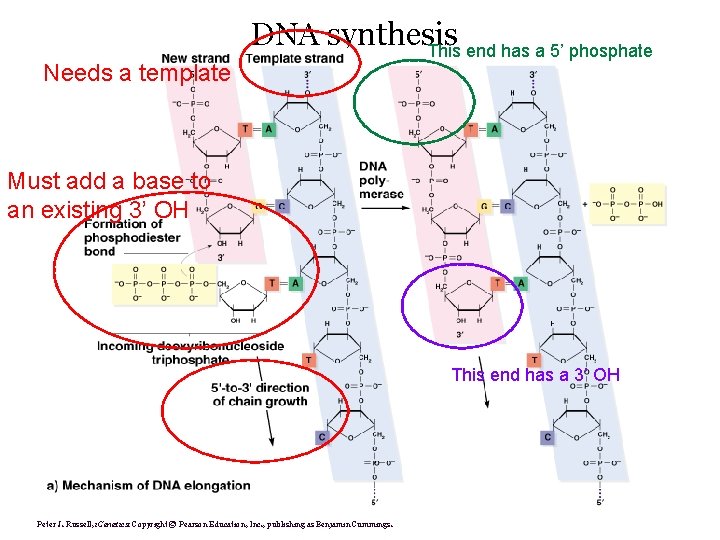
Needs a template DNA synthesis This end has a 5’ phosphate Must add a base to an existing 3’ OH This end has a 3’ OH Peter J. Russell, i. Genetics: Copyright © Pearson Education, Inc. , publishing as Benjamin Cummings.

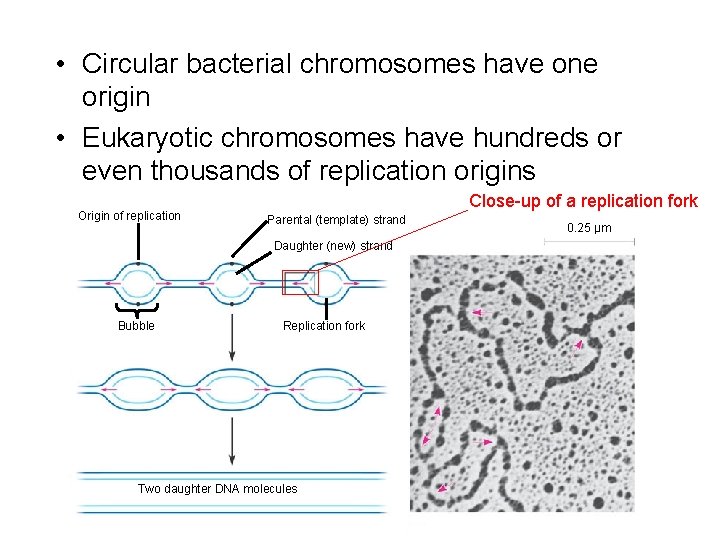
• Circular bacterial chromosomes have one origin • Eukaryotic chromosomes have hundreds or even thousands of replication origins Origin of replication Close-up of a replication fork Parental (template) strand Daughter (new) strand Bubble Replication fork Two daughter DNA molecules 0. 25 µm

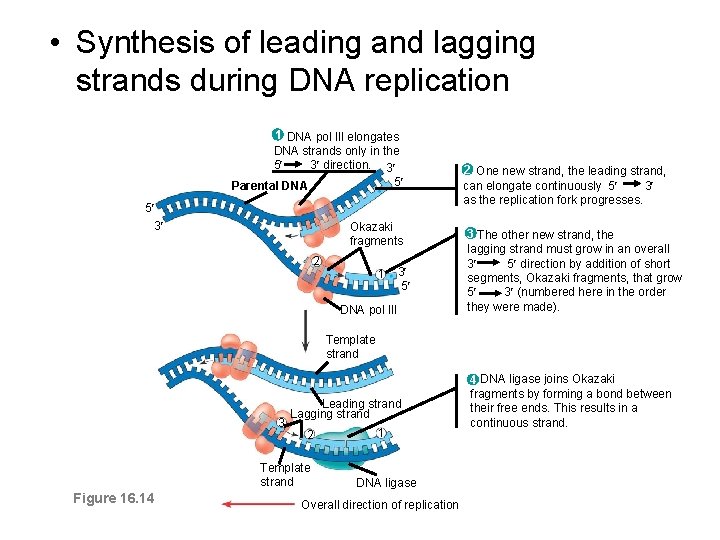
• Synthesis of leading and lagging strands during DNA replication 1 DNA pol Ill elongates DNA strands only in the 5 3 direction. 3 5 Parental DNA 5 3 Okazaki fragments 2 1 3 5 DNA pol III 2 One new strand, the leading strand, can elongate continuously 5 3 as the replication fork progresses. 3 The other new strand, the lagging strand must grow in an overall 3 5 direction by addition of short segments, Okazaki fragments, that grow 5 3 (numbered here in the order they were made). Template strand 3 Leading strand Lagging strand 2 Template strand Figure 16. 14 1 DNA ligase Overall direction of replication 4 DNA ligase joins Okazaki fragments by forming a bond between their free ends. This results in a continuous strand.

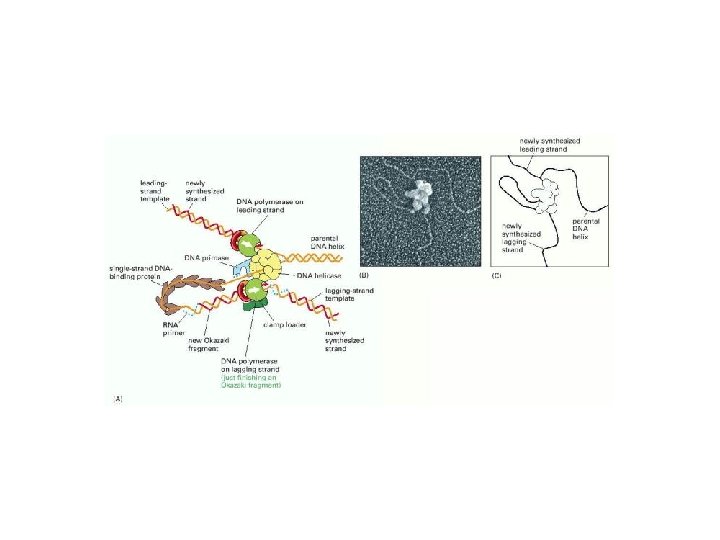
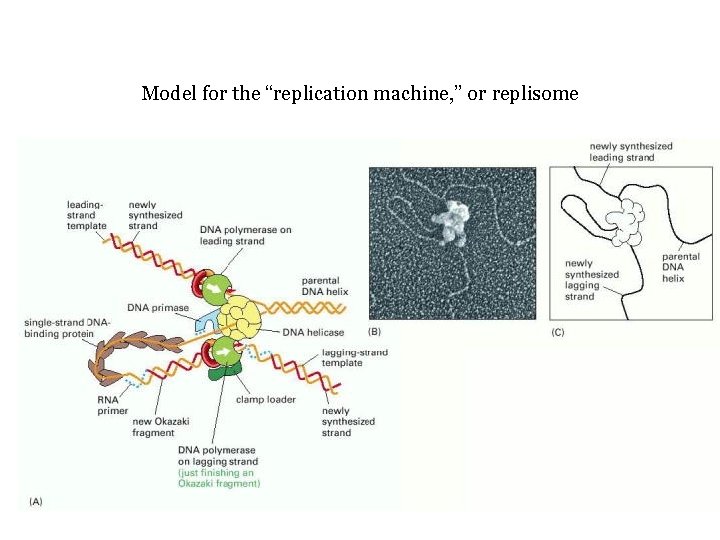
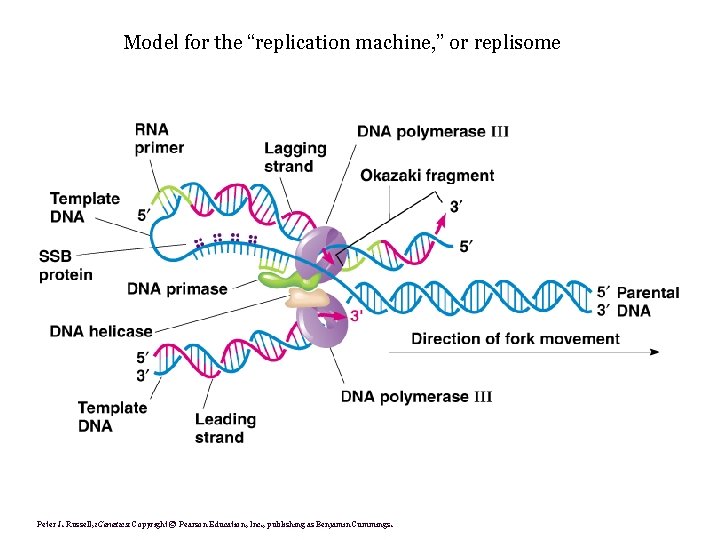
Model for the “replication machine, ” or replisome

Model for the “replication machine, ” or replisome Peter J. Russell, i. Genetics: Copyright © Pearson Education, Inc. , publishing as Benjamin Cummings.

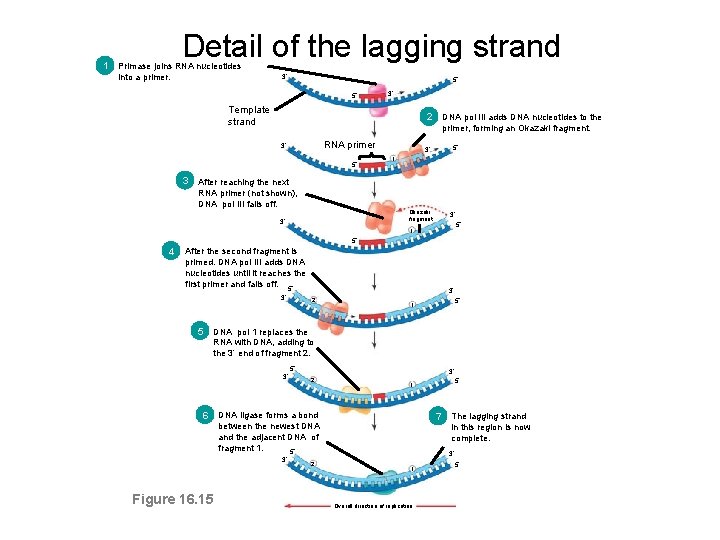
Detail of the lagging strand 1 Primase joins RNA nucleotides into a primer. 3 5 5 3 Template strand 2 DNA pol III adds DNA nucleotides to the primer, forming an Okazaki fragment. RNA primer 3 5 1 3 After reaching the next RNA primer (not shown), DNA pol III falls off. Okazaki fragment 3 3 5 1 5 4 After the second fragment is primed. DNA pol III adds DNA nucleotides until it reaches the first primer and falls off. 5 3 3 2 5 1 5 DNA pol 1 replaces the RNA with DNA, adding to the 3 end of fragment 2. 3 5 3 2 6 DNA ligase forms a bond 7 The lagging strand between the newest DNA and the adjacent DNA of fragment 1. 5 3 Figure 16. 15 2 5 1 in this region is now complete. 3 1 Overall direction of replication 5

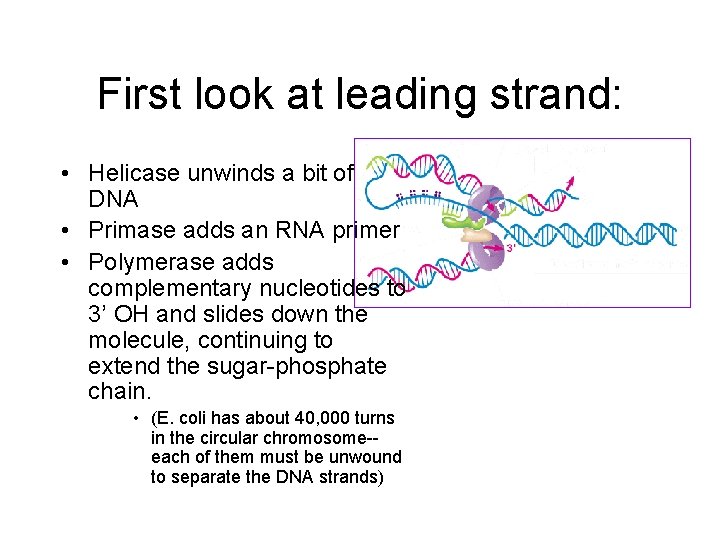
First look at leading strand: • Helicase unwinds a bit of DNA • Primase adds an RNA primer • Polymerase adds complementary nucleotides to 3’ OH and slides down the molecule, continuing to extend the sugar-phosphate chain. • (E. coli has about 40, 000 turns in the circular chromosome-each of them must be unwound to separate the DNA strands)

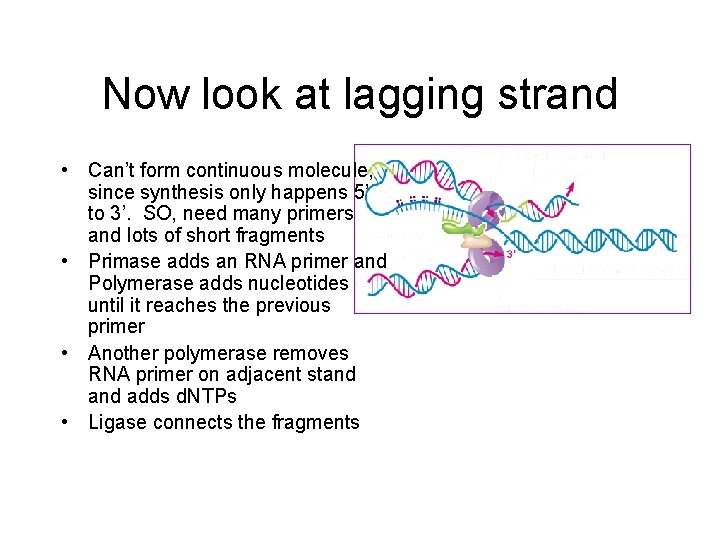
Now look at lagging strand • Can’t form continuous molecule, since synthesis only happens 5’ to 3’. SO, need many primers and lots of short fragments • Primase adds an RNA primer and Polymerase adds nucleotides until it reaches the previous primer • Another polymerase removes RNA primer on adjacent stand adds d. NTPs • Ligase connects the fragments

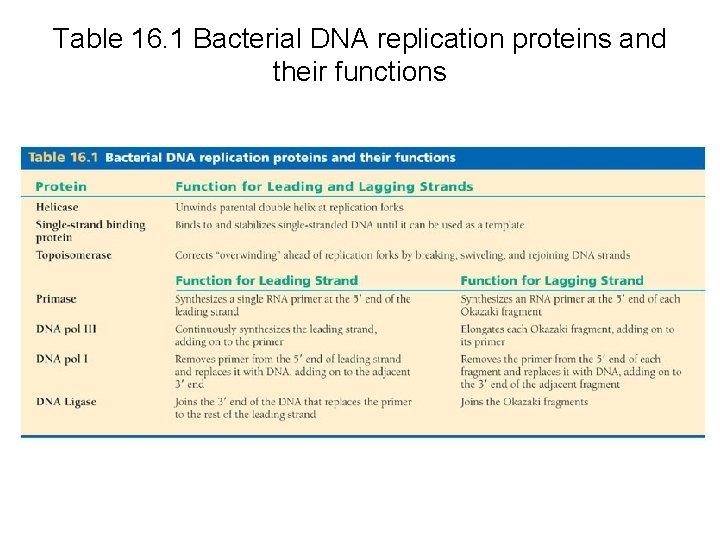
Table 16. 1 Bacterial DNA replication proteins and their functions

Try this at home: • Draw a replication bubble, label the bases on the template, primer, and newly synthesized DNA using the shorthand DNA notation. • Convince yourself that synthesis must go 5’ to 3’ and that replication must be semi-discontinuous.