Lecture 2 Identifying templates for protein modeling Sequence


















![PSI-BLAST is performed in five steps [1] Scan the protein database with a query PSI-BLAST is performed in five steps [1] Scan the protein database with a query](https://slidetodoc.com/presentation_image_h/27431925b7ada9a852c6e4ca0261b632/image-30.jpg)




![PSI-BLAST is performed in five steps [1] Scan the protein database with a query PSI-BLAST is performed in five steps [1] Scan the protein database with a query](https://slidetodoc.com/presentation_image_h/27431925b7ada9a852c6e4ca0261b632/image-36.jpg)

![PSI-BLAST is performed in five steps [1] Scan the protein database with a query PSI-BLAST is performed in five steps [1] Scan the protein database with a query](https://slidetodoc.com/presentation_image_h/27431925b7ada9a852c6e4ca0261b632/image-38.jpg)
- Slides: 39

Lecture 2 Identifying templates for protein modeling: Sequence alignment with BLAST and PSI-BLAST

Sources and additional information Images and other material in this presentation are taken from Bioinformatics and Functional Genomics third edition by Jonathan Pevsner, 2015 John Wiley & Sons, Inc. (http: //pevsnerlab. kennedykrieger. org/) The lecture follows closely the contents of chapter 4 of Pevsner book, which contains an in-depth discussion of the issues covered during the lecture. For additional material, please go to the book website: http: //www. bioinfbook. org

Sequence alignment with BLAST Webserver interface Aspects of the Algorithm Alignment strategies Detection of distant evolutionary relationships: PSI-BLAST

BLAST (Basic Local Alignment Search Tool) Scan large databases of sequences

Typical use • identifying orthologs and paralogs • discovering variants proteins • Exploring structure-function relations

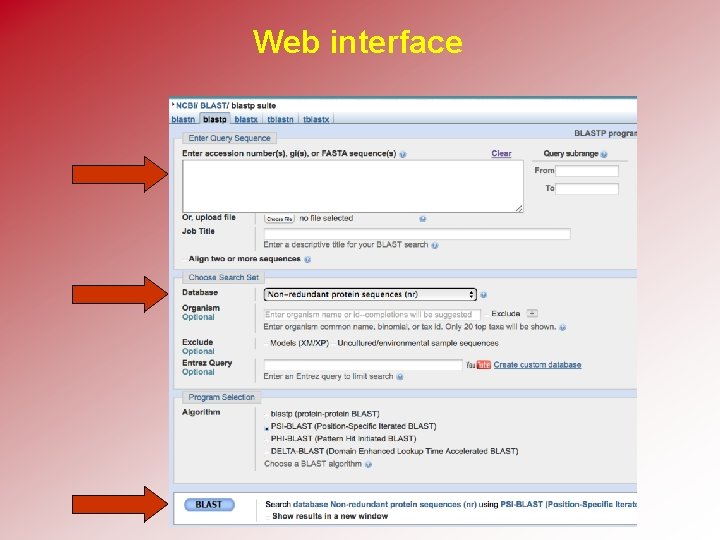
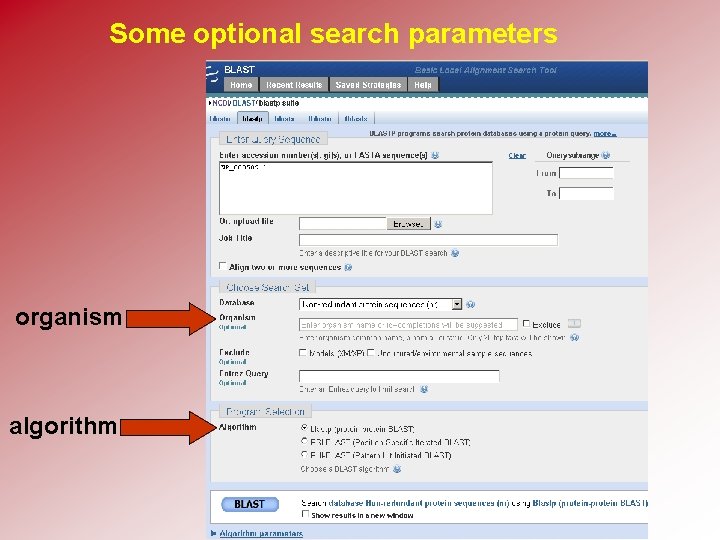
BLAST requires four choices • Choose the query sequence • Select the BLAST program • Choose a database • Select optional parameters

Web interface

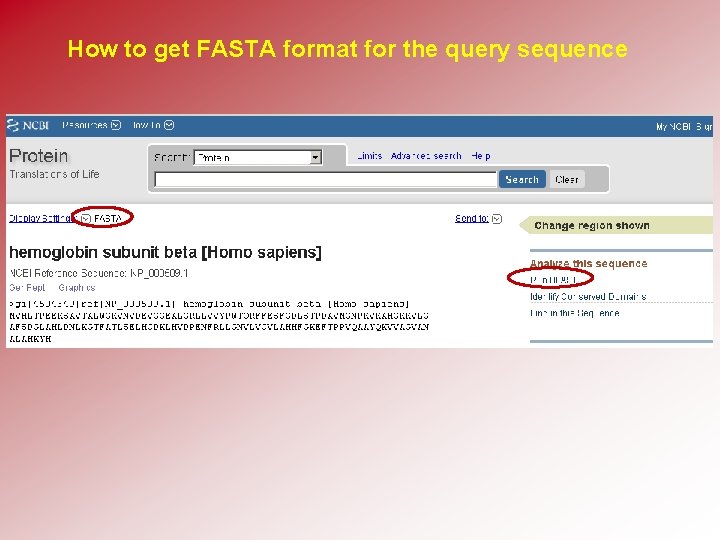
How to get FASTA format for the query sequence

Five distinct BLAST programs blastn (nucleotide BLAST) blastp (protein BLAST) blastx (translated BLAST) tblastn (translated BLAST) tblastx (translated BLAST)

Some optional search parameters organism algorithm

Why low complexity filter? (a) Query: human insulin NP_000198 Program: blastp Database: C. elegans Ref. Seq Default settings: Unfiltered (“composition-based statistics”)

Why low complexity filter? (d) Query: human insulin NP_000198 Program: blastp Database: C. elegans Ref. Seq Option: Filter low complexity regions Different bit score !

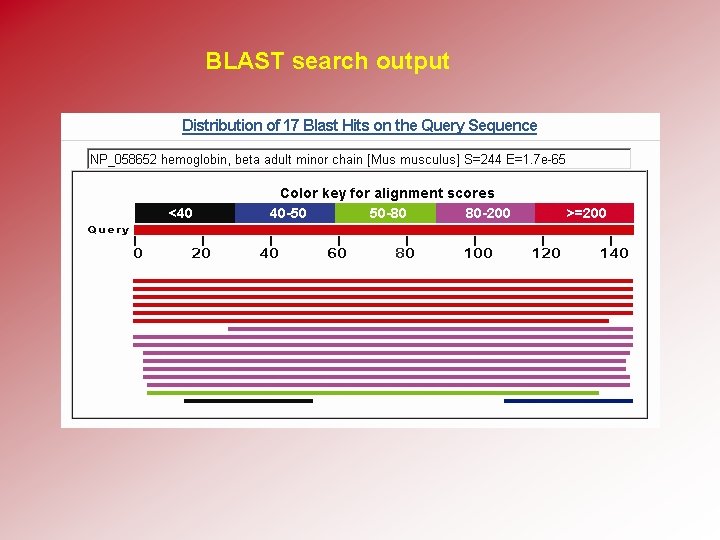
BLAST search output

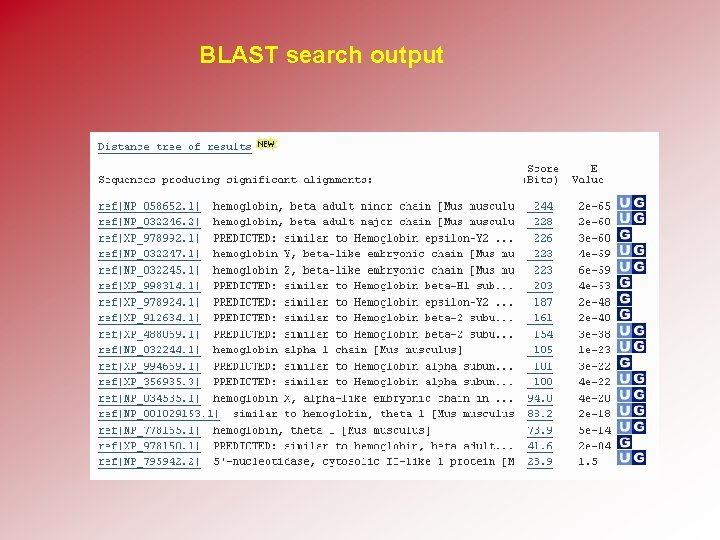
BLAST search output

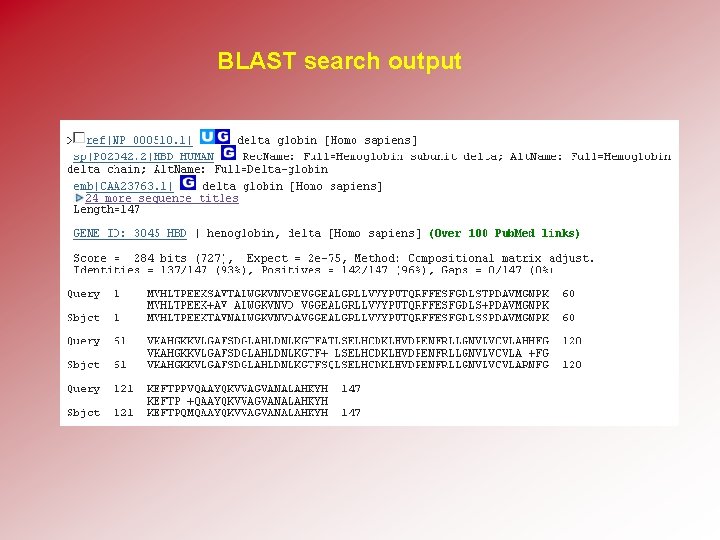
BLAST search output

Sequence alignment with BLAST Webserver interface Aspects of the Algorithm Alignment strategies Detection of distant evolutionary relationships: PSI-BLAST

BLAST: what kind of alignment? Global alignment (Needleman & Wunsch 1970): • Uses dynamic programming • Gaps are inserted so that the total lengths of both sequences are aligned (“global”).

BLAST: what kind of alignment? Local alignment (Smith & Waterman, 1980): • Just a portion of either sequence is aligned • Useful to find matching domains in two sequences. BLAST finds a local alignment through a heuristic approach

How the BLAST works: three phases Phase 1: compile a list of word pairs (w=3) above threshold T Example: for a human RBP query …FSGTWYA… (query word is in yellow) A list of words (w=3) is: FSG SGT GTW TWY WYA YSG TGT ATW SWY WFA FTG SVT GSW TWF WYS

Phase 1: compile a list of words (w=3) and score them according to BLOSUM matrices GTW 6, 5, 11 22 neighborhood GSW 6, 1, 11 18 word hits ATW 0, 5, 11 16 > threshold NTW 0, 5, 11 16 GTY 6, 5, 2 13 (T=11) GNW 10 neighborhood GAW 9 word hits < below threshold

BLAST second phase Phase 2: Scan the database to find matches for the compiled list.

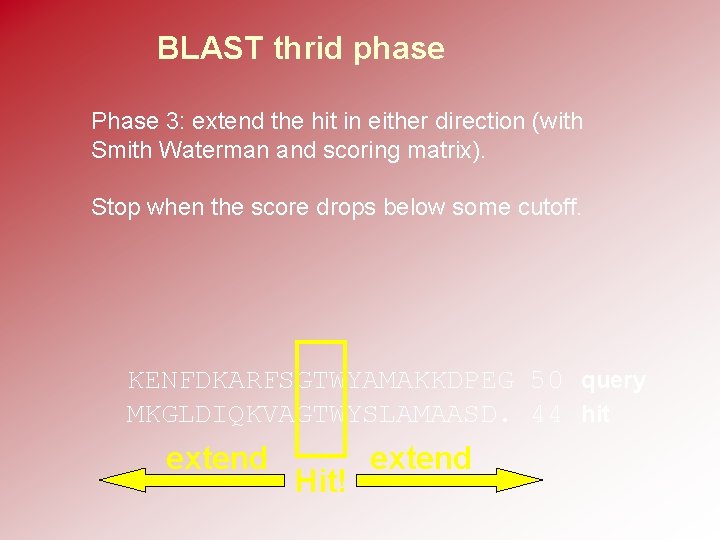
BLAST thrid phase Phase 3: extend the hit in either direction (with Smith Waterman and scoring matrix). Stop when the score drops below some cutoff. KENFDKARFSGTWYAMAKKDPEG 50 query MKGLDIQKVAGTWYSLAMAASD. 44 hit extend Hit! extend

How to interpret a BLAST search: expect value It is important to assess the statistical significance of search results. For local alignments (including BLAST search results), the statistics are well understood. The scores follow an extreme value distribution (EVD) rather than a normal distribution.

E = Kmn e-l. S E-value from extreme value distribution (number of highscoring segment pairs expected to occur with a score of at least S) S = the score m, n = the length of two sequences l, K = Karlin Altschul statistics (empirical)

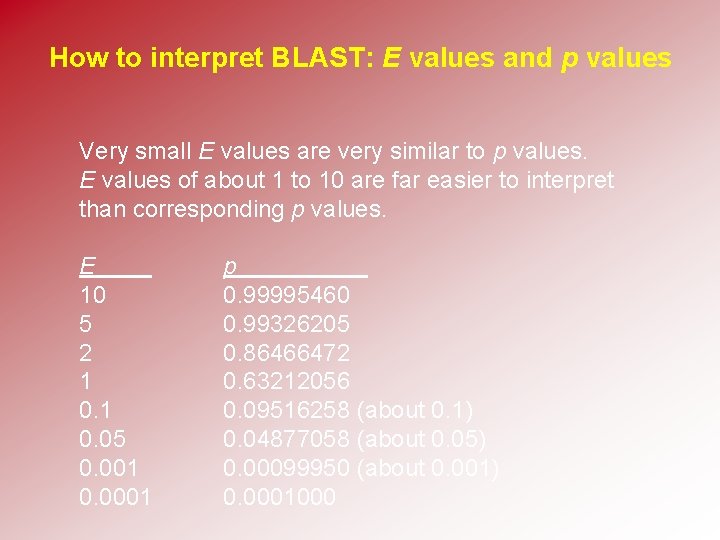
How to interpret BLAST: E values and p values Very small E values are very similar to p values. E values of about 1 to 10 are far easier to interpret than corresponding p values. E 10 5 2 1 0. 05 0. 001 0. 0001 p 0. 99995460 0. 99326205 0. 86466472 0. 63212056 0. 09516258 (about 0. 1) 0. 04877058 (about 0. 05) 0. 00099950 (about 0. 001) 0. 0001000

Sequence alignment with BLAST Webserver interface Aspects of the Algorithm Alignment strategies Detection of distant evolutionary relationships: PSI-BLAST

A real match might have E value > 1 Where do we stop? running BLAST with a putative hit as a query might help to establish a threshold

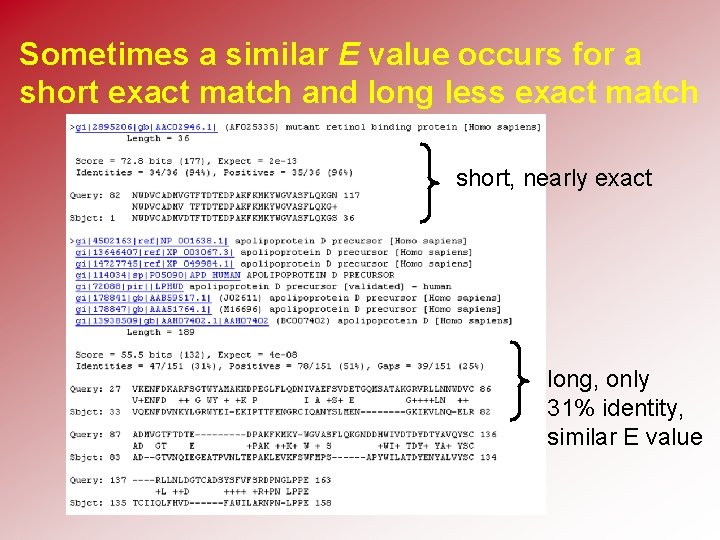
Sometimes a similar E value occurs for a short exact match and long less exact match short, nearly exact long, only 31% identity, similar E value

Sequence alignment with BLAST Webserver interface Aspects of the Algorithm Alignment strategies Detection of distant evolutionary relationships: PSI-BLAST
![PSIBLAST is performed in five steps 1 Scan the protein database with a query PSI-BLAST is performed in five steps [1] Scan the protein database with a query](https://slidetodoc.com/presentation_image_h/27431925b7ada9a852c6e4ca0261b632/image-30.jpg)
PSI-BLAST is performed in five steps [1] Scan the protein database with a query [2] PSI-BLAST uses the hits to generate a multiple sequence alignment. The latter is used to initialize a position-specific scoring matrix (PSSM)

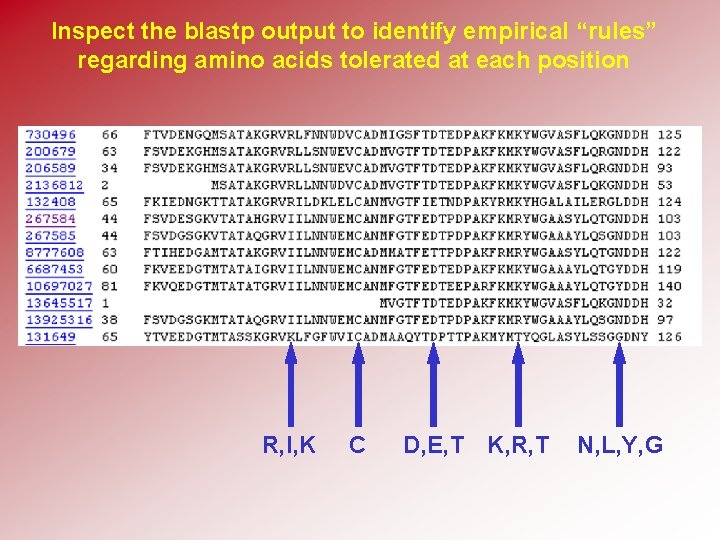
Inspect the blastp output to identify empirical “rules” regarding amino acids tolerated at each position R, I, K C D, E, T K, R, T N, L, Y, G

1 M 2 K 3 W 4 V 5 W 6 A 7 L 8 L 9 L 10 L 11 A 12 A 13 W 14 A 15 A 16 A. . . 37 S 38 G 39 T 40 W 41 Y 42 A A -1 -1 -3 0 -3 5 -2 -1 -1 -2 5 5 -2 3 2 4 R N D C Q E G H I L K M F -2 -2 -3 -2 -1 -2 -3 -2 1 2 -2 6 0 1 -4 2 4 -2 0 -3 -3 3 -2 -4 -3 -4 -5 -3 -2 -3 -2 1 20 amino acids -3 -3 -4 -1 -3 -3 -4 -4 3 1 -1 -3 -4 -5 -3 -2 -3 -2 1 -2 -2 -2 -1 -1 -1 0 -2 -2 -2 -1 -1 -3 -2 -4 -4 -1 -2 -3 -4 -3 2 0 -3 -3 -4 -1 -3 -3 -4 -3 2 2 -3 1 3 -3 -4 -4 -1 -2 -3 -4 -3 2 0 -2 -2 -2 -1 -1 -1 0 -2 -2 -2 -1 -1 -3 all the amino acids -3 -4 -4 -2 -2 -3 -4 -3 1 4 -3 2 1 from 1 to 4 the -2 -1 position -2 -1 -1 -2 -2 -1 -2 -3 -1 0 -1 -2 2 0 2 -1 -3 -3 0 -2 -3 end of your PSI-2 -1 -1 -1 3 -2 -2 -2 -1 -1 -3 P -3 -1 -4 -3 -4 -1 -3 -3 -1 -1 -1 W -2 -3 12 -3 -2 -2 -3 -3 7 -3 -3 -3 Y -1 -2 2 -1 2 -2 -1 0 -1 -1 -2 -2 0 -3 -2 -2 V 1 -3 -3 4 -3 0 1 3 2 1 0 0 0 -1 -2 -1 2 0 0 -3 -2 4 -1 -3 -2 -2 -1 4 1 -3 -2 0 -2 -3 -1 1 5 -3 -4 -3 -3 12 -3 -2 -2 2 -1 1 0 -3 -2 2 7 -2 -2 -4 0 -3 -1 0 BLAST query protein 0 -1 0 -4 -2 -2 -1 -5 -3 -2 -1 -3 -3 -1 0 -2 -1 -2 -2 -1 0 0 -1 -2 -3 -2 6 -2 -4 -4 -1 -2 -2 -1 -1 -3 -3 -2 -2 -3 2 -2 -1 -1 0 -2 -2 -2 0 -2 -1 -3 -2 -1 -2 -3 -1 -2 -1 -1 -3 -4 -2 1 3 -3 S -2 0 -3 -2 -3 1 -3 -2 -3 -3 1 1 -3 1 T -1 -1 -3 0 -1 -1 0 0 -2 -1 0 0

1 M 2 K 3 W 4 V 5 W 6 A 7 L 8 L 9 L 10 L 11 A 12 A 13 W 14 A 15 A 16 A. . . 37 S 38 G 39 T 40 W 41 Y 42 A A -1 -1 -3 0 -3 5 -2 -1 -1 -2 5 5 -2 3 2 4 R -2 1 -3 -3 -3 -2 -2 -2 -3 -2 -1 -2 N -2 0 -4 -3 -4 -2 -4 -3 -4 -4 -2 -2 -4 -1 0 -1 D -3 1 -5 -4 -5 -2 -4 -4 -2 -2 -4 -2 -1 -2 C -2 -4 -3 -1 -1 -2 -1 Q -1 2 -2 -3 -2 -1 -2 -3 -2 -2 -1 -1 -2 -1 E -2 4 -3 -3 -3 -1 -1 -3 -2 0 -1 G -3 -2 -3 -4 -3 0 -4 -4 0 0 -4 4 2 3 H -2 0 -3 -4 -3 -2 -3 -3 -2 -2 -3 -2 -1 -2 I 1 -3 -3 -2 2 2 -2 -2 1 -2 -3 -2 L 2 -3 -2 1 -2 -2 4 4 -2 -2 4 -2 -3 -2 K -2 3 -3 -3 -3 -1 -1 -3 -1 0 -1 M 6 -2 -2 1 -2 -1 2 2 -1 -1 2 -2 -2 -1 F 0 -4 1 -1 1 -3 0 0 -3 -3 1 -3 -3 -3 P -3 -1 -4 -3 -4 -1 -3 -3 -1 -1 -1 S -2 0 -3 -2 -3 1 -3 -2 -3 -3 1 1 -3 1 T -1 -1 -3 0 -1 -1 0 0 -2 -1 0 0 W -2 -3 12 -3 -2 -2 -3 -3 7 -3 -3 -3 Y -1 -2 2 -1 2 -2 -1 0 -1 -1 -2 -2 0 -3 -2 -2 V 1 -3 -3 4 -3 0 1 3 2 1 0 0 0 -1 -2 -1 2 0 0 -3 -2 4 -1 -3 -2 -2 0 -1 0 -4 -2 -2 -1 -5 -3 -2 -1 -3 -3 -1 0 -2 -1 -2 -2 -1 0 0 -1 -2 -3 -2 6 -2 -4 -4 -1 -2 -2 -1 -1 -3 -3 -2 -2 -3 2 -2 -1 -1 0 -2 -2 -2 0 -2 -1 -3 -2 -1 -2 -3 -1 -2 -1 -1 -3 -4 -2 1 3 -3 -1 4 1 -3 -2 0 -2 -3 -1 1 5 -3 -4 -3 -3 12 -3 -2 -2 2 -1 1 0 -3 -2 2 7 -2 -2 -4 0 -3 -1 0

1 M 2 K 3 W 4 V 5 W 6 A 7 L 8 L 9 L 10 L 11 A 12 A 13 W 14 A 15 A 16 A. . . 37 S 38 G 39 T 40 W 41 Y 42 A A -1 -1 -3 0 -3 5 -2 -1 -1 -2 5 5 -2 3 2 4 R -2 1 -3 -3 -3 -2 -2 -2 -3 -2 -1 -2 N -2 0 -4 -3 -4 -2 -4 -3 -4 -4 -2 -2 -4 -1 0 -1 D -3 1 -5 -4 -5 -2 -4 -4 -2 -2 -4 -2 -1 -2 C Q E G H I L K M -2 -1 -2 -3 -2 1 2 -2 6 -4 2 4 -2 0 -3 -3 3 -2 -3 -3 -2 -1 -3 -3 -4 -4 3 1 -3 -2 -3 -2 -1 -1 -1 0 -2 -2 -2 -1 -1 -1 -2 -3 -4 -3 2 -1 -3 -3 -4 -3 2 2 -3 1 -1 -2 -3 -4 a-3 2 4 -3 2 note that given -1 -2 -3 -4 -3 2 amino -1 -1 -1 acid 0 -2(such -2 -2 as -1 -1 -1 0 in-2 -2 -2 -1 -1 alanine) your query -2 -2 -3 -4 -3 1 4 -3 2 protein -1 -1 -2 can 4 -2 receive -2 -2 -1 -2 -2 2 0 2 scores -1 -3 -3 different for 0 -2 -1 -1 -1 3 -2 -2 -2 -1 -1 2 0 0 -3 -2 4 -1 -3 -2 -2 0 -1 0 -4 -2 -2 -1 -3 -2 -4 -1 -2 -5 -3 -2 -3 -2 1 -3 -3 -2 -2 -3 2 -2 -1 3 -2 -1 -1 -1 0 -2 -2 -2 -1 -1 -3 matching alanine— -1 0 0 0 -1 -3 0 -2 depending on-2 the -3 -2 -2 6 -2 -4 -4 -2 -3 position in -2 the-1 protein -1 -1 -1 -2 -1 -1 -1 F 0 -4 1 -1 1 -3 0 0 -3 -3 1 -3 -3 -3 P -3 -1 -4 -3 -4 -1 -3 -3 -1 -1 -1 S -2 0 -3 -2 -3 1 -3 -2 -3 -3 1 1 -3 1 T -1 -1 -3 0 -1 -1 0 0 -2 -1 0 0 W -2 -3 12 -3 -2 -2 -3 -3 7 -3 -3 -3 Y -1 -2 2 -1 2 -2 -1 0 -1 -1 -2 -2 0 -3 -2 -2 V 1 -3 -3 4 -3 0 1 3 2 1 0 0 0 -1 -2 -1 -1 4 1 -3 -2 0 -2 -3 -1 1 5 -3 -4 -3 -3 12 -3 -2 -2 2 -1 1 0 -3 -2 2 7 -2 -2 -4 0 -3 -1 0

1 M 2 K 3 W 4 V 5 W 6 A 7 L 8 L 9 L 10 L 11 A 12 A 13 W 14 A 15 A 16 A. . . 37 S 38 G 39 T 40 W 41 Y 42 A A -1 -1 -3 0 -3 5 -2 -1 -1 -2 5 5 -2 3 2 4 R -2 1 -3 -3 -3 -2 -2 -2 -3 -2 -1 -2 N -2 0 -4 -3 -4 -2 -4 -3 -4 -4 -2 -2 -4 -1 0 -1 D -3 1 -5 -4 -5 -2 -4 -4 -2 -2 -4 -2 -1 -2 C -2 -4 -3 -1 -1 -2 -1 Q E G H I L K M -1 -2 -3 -2 1 2 -2 6 2 4 -2 0 -3 -3 3 -2 -2 -3 -3 -4 -4 3 1 -2 -3 -3 -2 -1 -1 0 -2 -2 -2 -1 -1 -2 -3 -4 -3 2 -3 -3 -4 -3 2 2 -3 1 -2 -3 that -4 -3 2 note a given -2 -3 -4 -3 2 amino acid (such as-1 -1 -1 0 -2 -2 -2 -1 -1 -1 0 -2 -2 -1 -1 tryptophan) in -2 your -2 -3 -4 -3 1 4 -3 2 query -1 -2 protein 4 -2 -2 can -2 -1 -2 2 0 2 different -1 -3 -3 0 -2 receive -1 -1 3 -2 -2 -2 -1 -1 F 0 -4 1 -1 1 -3 0 0 -3 -3 1 -3 -3 -3 2 0 0 -3 -2 4 -1 -3 -2 -2 0 -1 0 -4 -2 -2 -1 -5 -3 -3 -2 -2 -3 2 -2 -1 3 -2 -1 -1 -1 0 -2 -2 -2 -1 -1 -3 scores for matching -1 tryptophan— 0 0 0 -1 -2 -3 0 -2 -3 -3 -2 -2 6 -2 -4 -4 -2 -3 -4 the-1 -1 -2 -1 depending -1 -1 -2 -2 on -1 -1 -3 position -2 -3 -3 in -3 the -3 -2 1 protein P -3 -1 -4 -3 -4 -1 -3 -3 -1 -1 -1 S -2 0 -3 -2 -3 1 -3 -2 -3 -3 1 1 -3 1 T -1 -1 -3 0 -1 -1 0 0 -2 -1 0 0 W -2 -3 12 -3 -2 -2 -3 -3 7 -3 -3 -3 Y -1 -2 2 -1 2 -2 -1 0 -1 -1 -2 -2 0 -3 -2 -2 V 1 -3 -3 4 -3 0 1 3 2 1 0 0 0 -1 -2 -1 -1 4 1 -3 -2 0 -2 -3 -1 1 5 -3 -4 -3 -3 12 -3 -2 -2 2 -1 1 0 -3 -2 2 7 -2 -2 -4 0 -3 -1 0
![PSIBLAST is performed in five steps 1 Scan the protein database with a query PSI-BLAST is performed in five steps [1] Scan the protein database with a query](https://slidetodoc.com/presentation_image_h/27431925b7ada9a852c6e4ca0261b632/image-36.jpg)
PSI-BLAST is performed in five steps [1] Scan the protein database with a query [2] PSI-BLAST uses the hits to generate a multiple sequence alignment. The latter is used to initialize a position-specific scoring matrix (PSSM) [3] The PSSM is used to score the alignments of the query with the database [4] Statistical significance (E values) are re-estimated on the basis of the new raw scores (from the PSSM)

Note the new entries: some hits bacame statistically significant with the PSSM
![PSIBLAST is performed in five steps 1 Scan the protein database with a query PSI-BLAST is performed in five steps [1] Scan the protein database with a query](https://slidetodoc.com/presentation_image_h/27431925b7ada9a852c6e4ca0261b632/image-38.jpg)
PSI-BLAST is performed in five steps [1] Scan the protein database with a query [2] PSI-BLAST uses the hits to generate a multiple sequence alignment. The latter is used to initialize a position-specific scoring matrix (PSSM) [3] The PSSM is used to score the alignments of the query with the database [4] Statistical significance (E values) are re-estimated on the basis of the new raw scores (from the PSSM) [5] Iterate through [3] and [4 until convergence (only in principle, in practice two or three times)

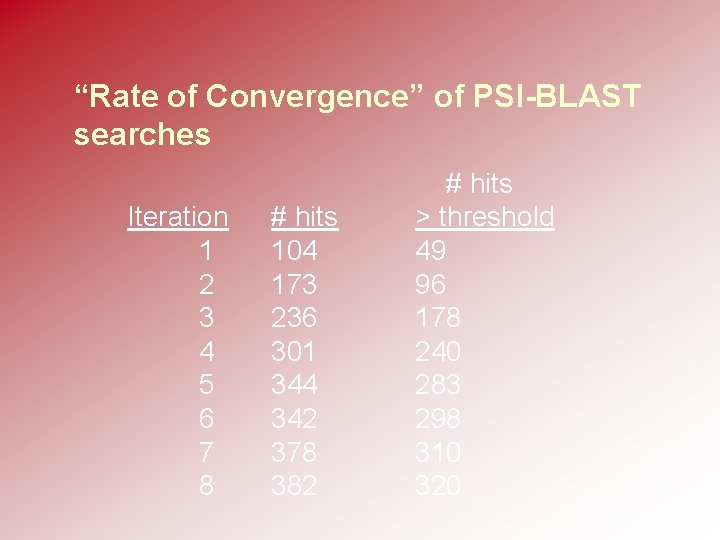
“Rate of Convergence” of PSI-BLAST searches Iteration 1 2 3 4 5 6 7 8 # hits 104 173 236 301 344 342 378 382 # hits > threshold 49 96 178 240 283 298 310 320