Lecture 10 Evolution Classification contd Species Classification Phenetic










- Slides: 24

Lecture 10: Evolution & Classification cont’d Species Classification: • Phenetic: physical attributes, numerical taxonomy • Cladistic (Phylogenetic): evolutionary relationships • Evolutionary: synthesis of the two Reflect Philosophical Differences

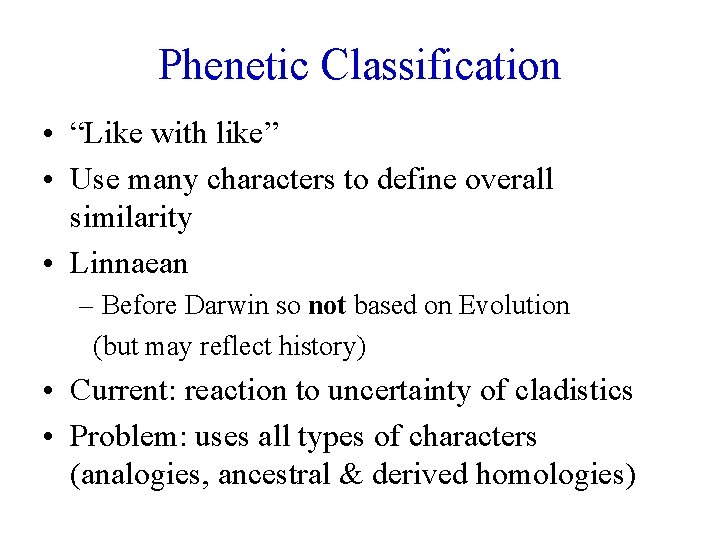
Phenetic Classification • “Like with like” • Use many characters to define overall similarity • Linnaean – Before Darwin so not based on Evolution (but may reflect history) • Current: reaction to uncertainty of cladistics • Problem: uses all types of characters (analogies, ancestral & derived homologies)

Steps 1) identify taxa to be considered 2) choose characters (independent, “unit”) 3) construct character matrix for each taxon: 4) use mathematical formula to describe degree of similarity for each taxon: e. g. simple matching coefficient # matches S = total # of characters

5) construct matrix with pairwise S values 6) use clustering technique to produce a dendrogram e. g. UPGMA (Unweighted Pair Group Method with Arithmetic Averaging) or Neighbour-joining Unweighted/Equal weighting = all characters given equal consideration unweighting is a type of weighting! may introduce bias…. .

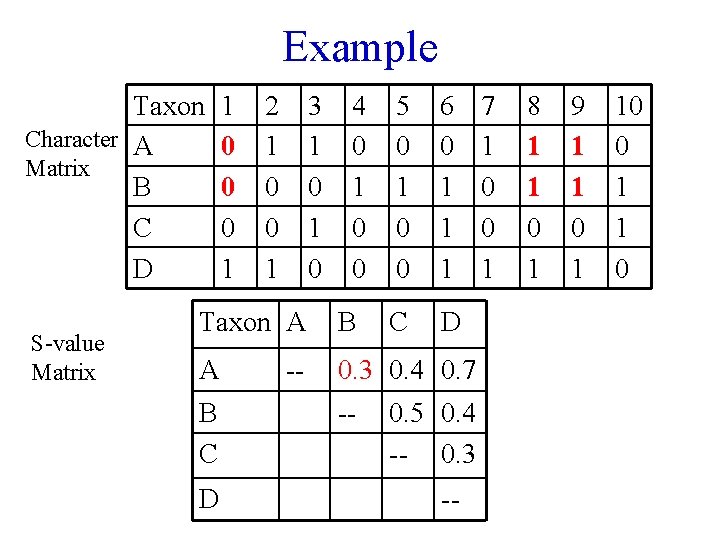
Example Taxon Character A Matrix B C D S-value Matrix 1 0 0 0 1 2 1 0 0 1 3 1 0 4 0 1 0 0 5 0 1 0 0 6 0 1 1 1 C D Taxon A B A 0. 3 0. 4 0. 7 B C D -- -- 0. 5 0. 4 -- 0. 3 -- 7 1 0 0 1 8 1 1 0 1 9 1 1 0 1 10 0 1 1 0

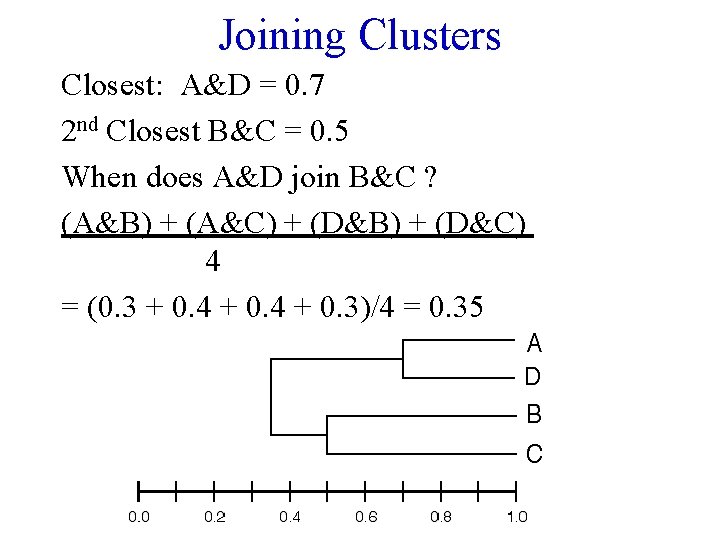
Joining Clusters Closest: A&D = 0. 7 2 nd Closest B&C = 0. 5 When does A&D join B&C ? (A&B) + (A&C) + (D&B) + (D&C) 4 = (0. 3 + 0. 4 + 0. 3)/4 = 0. 35

Problems • Different methods or characters = different dendrograms • If used all characteristics would = natural classification (Impossible!) • dendrogram = phylogeny if differences between taxa proportional to time elapsed since common ancestor

Unfortunately… 1. Mosaic Evolution: differences in rate of change of characters in a lineage 2. Homoplasy: shared characters not in common ancestor (analogy)

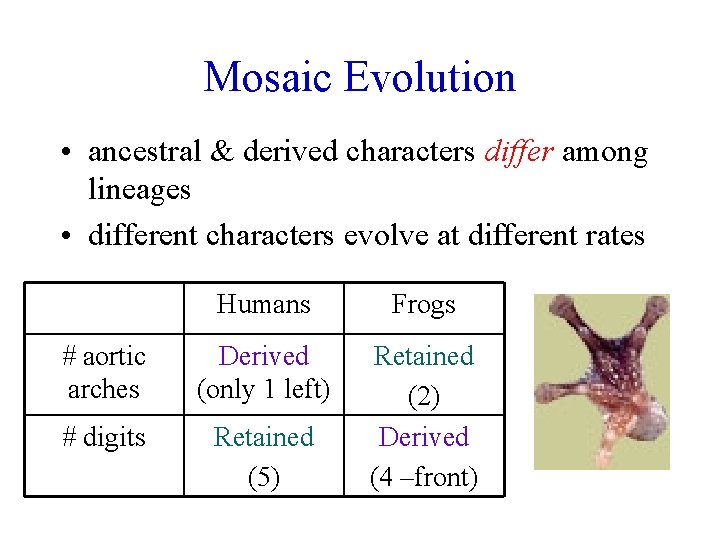
Mosaic Evolution • ancestral & derived characters differ among lineages • different characters evolve at different rates Humans Frogs # aortic arches Derived (only 1 left) # digits Retained (5) Retained (2) Derived (4 –front)

Why retained? A) Developmental Canalization • Character change requires change in developmental program (rare) B) General Adaptations • Useful in large number of ecological contexts e. g. Rodentia - incisors conserved - legs evolved rapidly

Homoplasy # characters used, chance of homoplasy A. Convergent Evolution • Similar phenotypic response to similar ecological conditions • Different developmental pathways

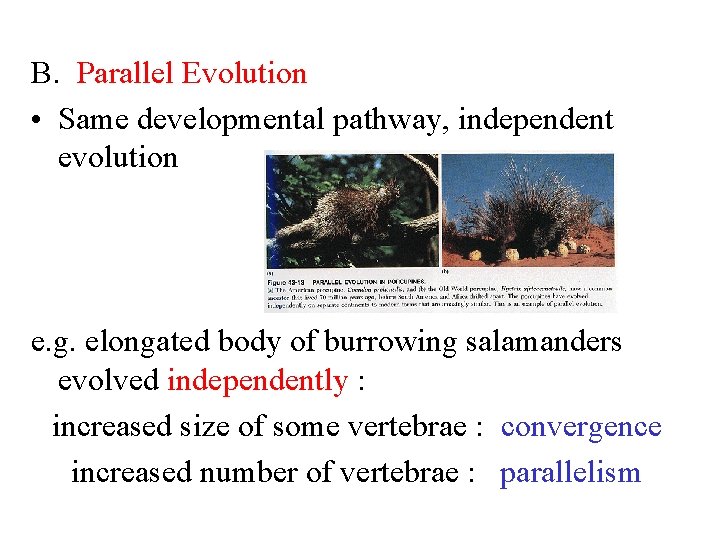
B. Parallel Evolution • Same developmental pathway, independent evolution e. g. elongated body of burrowing salamanders evolved independently : increased size of some vertebrae : convergence increased number of vertebrae : parallelism

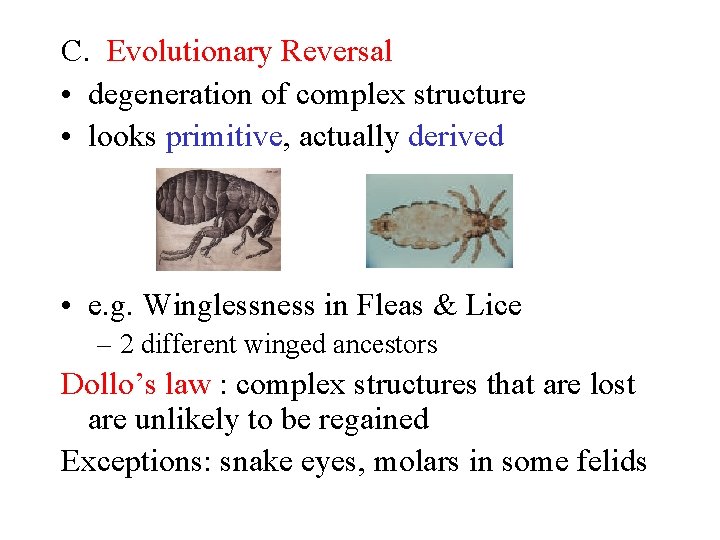
C. Evolutionary Reversal • degeneration of complex structure • looks primitive, actually derived • e. g. Winglessness in Fleas & Lice – 2 different winged ancestors Dollo’s law : complex structures that are lost are unlikely to be regained Exceptions: snake eyes, molars in some felids

Frogs with Teeth? • Reversals & Parallelism common because of potentialities (bias) of developmental systems • Frogs lost teeth in lower jaw in the Jurassic • Teeth can be expt’lly induced • Gastrotheca guentheri – re-evolved true teeth

• Homoplasy & variation in rate of character change = phenetic classification that may not show evolutionary history • Can get : monophyletic, paraphyletic, polyphyletic groups b/c use all character types

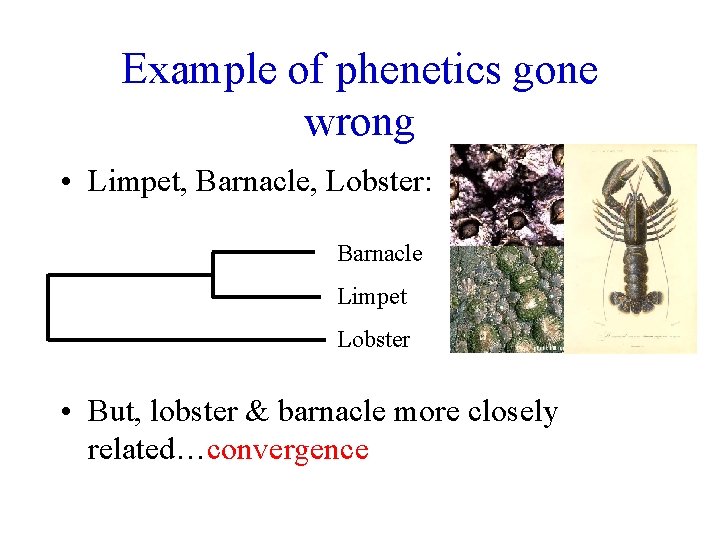
Example of phenetics gone wrong • Limpet, Barnacle, Lobster: Barnacle Limpet Lobster • But, lobster & barnacle more closely related…convergence

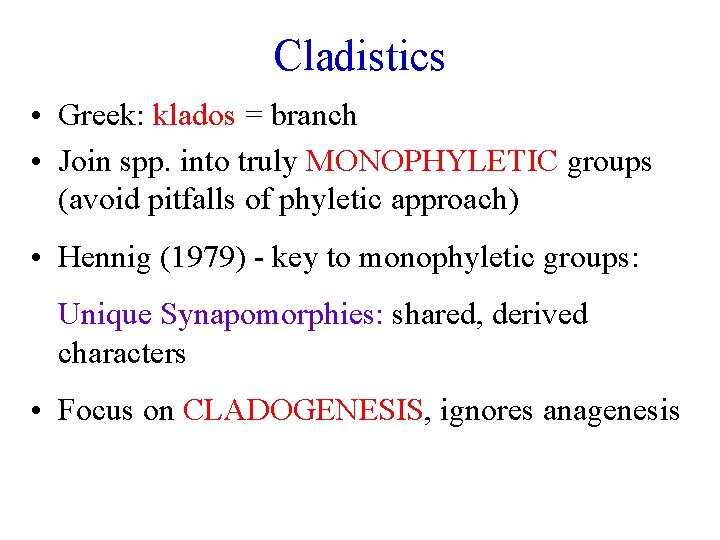
Cladistics • Greek: klados = branch • Join spp. into truly MONOPHYLETIC groups (avoid pitfalls of phyletic approach) • Hennig (1979) - key to monophyletic groups: Unique Synapomorphies: shared, derived characters • Focus on CLADOGENESIS, ignores anagenesis

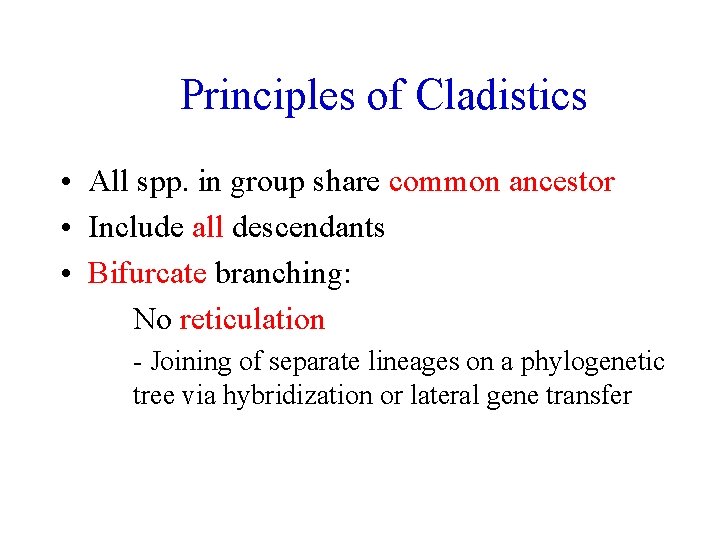
Principles of Cladistics • All spp. in group share common ancestor • Include all descendants • Bifurcate branching: No reticulation - Joining of separate lineages on a phylogenetic tree via hybridization or lateral gene transfer

Ancestral traits Criteria to determine primitiveness: • Presence in fossils • Commonness across taxa • Early appearance in phylogeny • Presence in outgroup

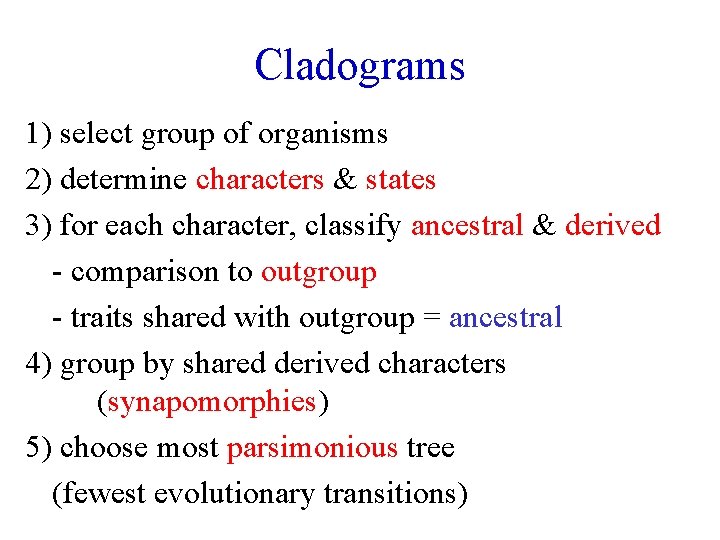
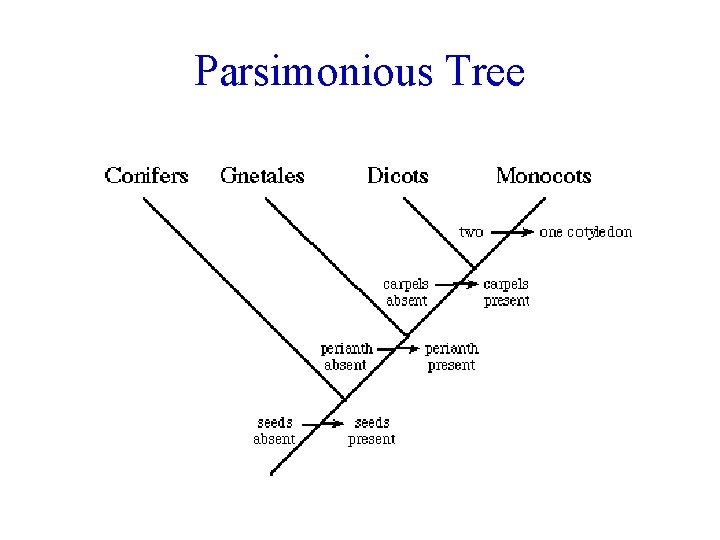
Cladograms 1) select group of organisms 2) determine characters & states 3) for each character, classify ancestral & derived - comparison to outgroup - traits shared with outgroup = ancestral 4) group by shared derived characters (synapomorphies) 5) choose most parsimonious tree (fewest evolutionary transitions)

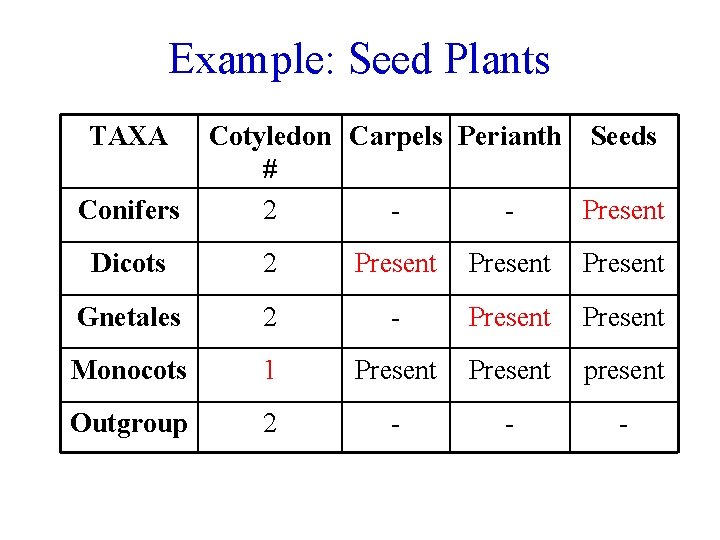
Example: Seed Plants TAXA Conifers Cotyledon Carpels Perianth Seeds # 2 Present Dicots 2 Present Gnetales 2 - Present Monocots 1 Present present Outgroup 2 - - -

Parsimonious Tree

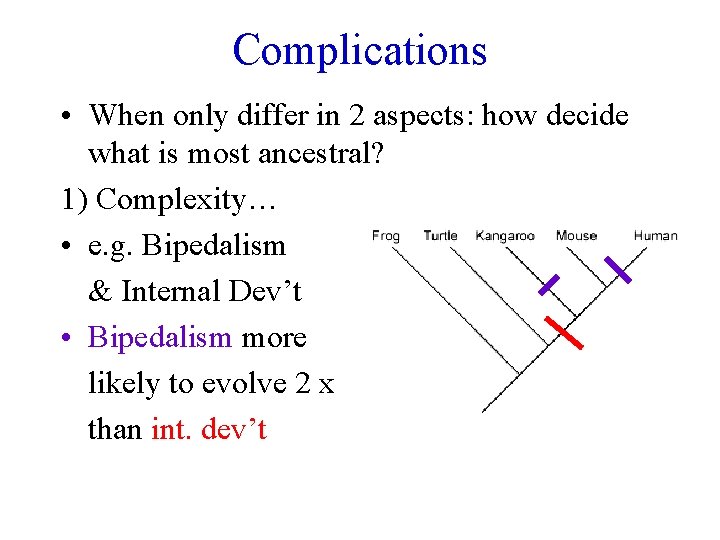
Complications • When only differ in 2 aspects: how decide what is most ancestral? 1) Complexity… • e. g. Bipedalism & Internal Dev’t • Bipedalism more likely to evolve 2 x than int. dev’t

2) SINEs & LINEs • • Short & Long Interspersed Elements Parasitic DNA sequences Can use as phylogenetic characters Insertion rare, unlikely to get same insertions from different events • Reversal detectable because lose part of host genome too • Homoplasy unlikely, reliable characters • Helped to determine place of whales in artiodactyla